Characterization of two novel polyomaviruses of birds by using multiply primed rolling-circle amplification of their genomes
- PMID: 16537620
- PMCID: PMC1440385
- DOI: 10.1128/JVI.80.7.3523-3531.2006
Characterization of two novel polyomaviruses of birds by using multiply primed rolling-circle amplification of their genomes
Abstract
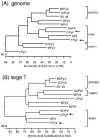
Polyomaviruses are small nonenveloped particles with a circular double-stranded genome, approximately 5 kbp in size. The mammalian polyomaviruses mainly cause persistent subclinical infections in their natural nonimmunocompromised hosts. In contrast, the polyomaviruses of birds--avian polyomavirus (APV) and goose hemorrhagic polyomavirus (GHPV)--are the primary agents of acute and chronic disease with high mortality rates in young birds. Screening of field samples of diseased birds by consensus PCR revealed the presence of two novel polyomaviruses in the liver of an Eurasian bullfinch (Pyrrhula pyrrhula griseiventris) and in the spleen of a Eurasian jackdaw (Corvus monedula), tentatively designated as finch polyomavirus (FPyV) and crow polyomavirus (CPyV), respectively. The genomes of the viruses were amplified by using multiply primed rolling-circle amplification and cloned. Analysis of the FPyV and CPyV genome sequences revealed a close relationship to APV and GHPV, indicating the existence of a distinct avian group among the polyomaviruses. The main characteristics of this group are (i) involvement in fatal disease, (ii) the existence of an additional open reading frame in the 5' region of the late mRNAs, and (iii) a different manner of DNA binding of the large tumor antigen compared to that of the mammalian polyomaviruses.
Figures

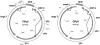


Similar articles
-
Whole-genome characterization of a novel polyomavirus detected in fatally diseased canary birds.J Gen Virol. 2010 Dec;91(Pt 12):3016-22. doi: 10.1099/vir.0.023549-0. Epub 2010 Aug 25. J Gen Virol. 2010. PMID: 20797969
-
Molecular characterization of avian polyomavirus isolated from psittacine birds based on the whole genome sequence analysis.Vet Microbiol. 2009 Jul 2;138(1-2):69-77. doi: 10.1016/j.vetmic.2009.03.007. Epub 2009 Mar 13. Vet Microbiol. 2009. PMID: 19345024
-
Genome analysis of the new human polyomaviruses.Rev Med Virol. 2012 Nov;22(6):354-77. doi: 10.1002/rmv.1711. Epub 2012 Mar 28. Rev Med Virol. 2012. PMID: 22461085 Review.
-
[Detection of DNA of the finch polyomavirus in diseases of various types of birds in the order Passeriformes].Berl Munch Tierarztl Wochenschr. 2007 Mar-Apr;120(3-4):113-9. Berl Munch Tierarztl Wochenschr. 2007. PMID: 17416133 German.
-
Genome analysis of non-human primate polyomaviruses.Infect Genet Evol. 2014 Aug;26:283-94. doi: 10.1016/j.meegid.2014.05.030. Epub 2014 Jun 14. Infect Genet Evol. 2014. PMID: 24933462 Review.
Cited by
-
Fecal, oral, blood and skin virome of laboratory rabbits.Arch Virol. 2020 Dec;165(12):2847-2856. doi: 10.1007/s00705-020-04808-y. Epub 2020 Oct 9. Arch Virol. 2020. PMID: 33034764 Free PMC article.
-
Complete Genome Sequence of a Novel Avian Polyomavirus Isolated from Gouldian Finch.Genome Announc. 2015 Sep 17;3(5):e01001-15. doi: 10.1128/genomeA.01001-15. Genome Announc. 2015. PMID: 26383660 Free PMC article.
-
Viral metagenomics.Rev Med Virol. 2007 Mar-Apr;17(2):115-31. doi: 10.1002/rmv.532. Rev Med Virol. 2007. PMID: 17295196 Free PMC article. Review.
-
Codon usage patterns of LT-Ag genes in polyomaviruses from different host species.Virol J. 2019 Nov 14;16(1):137. doi: 10.1186/s12985-019-1245-2. Virol J. 2019. PMID: 31727090 Free PMC article.
-
Possible cross-species transmission of circoviruses and cycloviruses among farm animals.J Gen Virol. 2011 Apr;92(Pt 4):768-72. doi: 10.1099/vir.0.028704-0. Epub 2010 Dec 22. J Gen Virol. 2011. PMID: 21177928 Free PMC article.
References
-
- Arnal, M. C., M. Revilla, M. Marco, U. Höfle, R. Mateo, J. M. Peiró, C. de Frutos, M. A. Jiménez, A. Sánchez, and D. Fernández de Luco. 2005. Brote de salmonelosis septicémica en estorninos pintos Sturnus vulgaris, p.106. Abstr. 17th Meet. Spanish Vet. Pathol. Soc., Jarandilla de la Vera, Cáceres, Spain, 14-16 July 2005.
-
- Carstans, E. B., M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Polyomaviridae, p. 241-246. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and E. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
-
- Cole, C. N., and S. D. Conzen. 2001. Polyomaviridae: the viruses and their replication, p. 2141-2174. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

