Enzymatic aminoacylation of tRNA with unnatural amino acids
- PMID: 16537388
- PMCID: PMC1450175
- DOI: 10.1073/pnas.0509219103
Enzymatic aminoacylation of tRNA with unnatural amino acids
Abstract
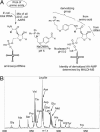
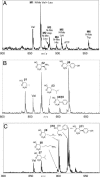
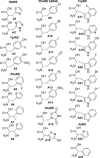
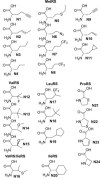
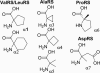
The biochemical flexibility of the cellular translation apparatus offers, in principle, a simple route to the synthesis of drug-like modified peptides and novel biopolymers. However, only approximately 75 unnatural building blocks are known to be fully compatible with enzymatic tRNA acylation and subsequent ribosomal synthesis of modified peptides. Although the translation system can reject substrate analogs at several steps along the pathway to peptide synthesis, much of the specificity resides at the level of the aminoacyl-tRNA synthetase (AARS) enzymes that are responsible for charging tRNAs with amino acids. We have developed an AARS assay based on mass spectrometry that can be used to rapidly identify unnatural monomers that can be enzymatically charged onto tRNA. By using this assay, we have found 59 previously unknown AARS substrates. These include numerous side-chain analogs with useful functional properties. Remarkably, many beta-amino acids, N-methyl amino acids, and alpha,alpha-disubstituted amino acids are also AARS substrates. These previously unidentified AARS substrates will be useful in studies of the specificity of subsequent steps in translation and may significantly expand the number of analogs that can be used for the ribosomal synthesis of modified peptides.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

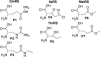
Similar articles
-
An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides.PLoS One. 2007 Oct 3;2(10):e972. doi: 10.1371/journal.pone.0000972. PLoS One. 2007. PMID: 17912351 Free PMC article.
-
Enzymatic tRNA acylation by acid and alpha-hydroxy acid analogues of amino acids.Biochemistry. 2008 Jan 8;47(1):301-7. doi: 10.1021/bi701456c. Epub 2007 Dec 8. Biochemistry. 2008. PMID: 18067322
-
tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.Nucleic Acids Res. 2024 Jan 25;52(2):513-524. doi: 10.1093/nar/gkad1188. Nucleic Acids Res. 2024. PMID: 38100361 Free PMC article.
-
Aminoacyl-tRNA Synthetases and tRNAs for an Expanded Genetic Code: What Makes them Orthogonal?Int J Mol Sci. 2019 Apr 19;20(8):1929. doi: 10.3390/ijms20081929. Int J Mol Sci. 2019. PMID: 31010123 Free PMC article. Review.
-
Synthetic and editing reactions of aminoacyl-tRNA synthetases using cognate and non-cognate amino acid substrates.Methods. 2017 Jan 15;113:13-26. doi: 10.1016/j.ymeth.2016.09.015. Epub 2016 Oct 3. Methods. 2017. PMID: 27713080 Review.
Cited by
-
Translational incorporation of modified phenylalanines and tyrosines during cell-free protein synthesis.RSC Adv. 2020 Mar 18;10(19):11013-11023. doi: 10.1039/d0ra00655f. eCollection 2020 Mar 16. RSC Adv. 2020. PMID: 35495348 Free PMC article.
-
In vitro genetic code reprogramming and expansion to study protein function and discover macrocyclic peptide ligands.Curr Opin Chem Biol. 2018 Oct;46:172-179. doi: 10.1016/j.cbpa.2018.07.013. Epub 2018 Aug 2. Curr Opin Chem Biol. 2018. PMID: 30077877 Free PMC article. Review.
-
Genetically encoded libraries of nonstandard peptides.J Nucleic Acids. 2012;2012:713510. doi: 10.1155/2012/713510. Epub 2012 Oct 14. J Nucleic Acids. 2012. PMID: 23097693 Free PMC article.
-
Foldamers as versatile frameworks for the design and evolution of function.Nat Chem Biol. 2007 May;3(5):252-62. doi: 10.1038/nchembio876. Nat Chem Biol. 2007. PMID: 17438550 Free PMC article. Review.
-
Biochemistry of Aminoacyl tRNA Synthetase and tRNAs and Their Engineering for Cell-Free and Synthetic Cell Applications.Front Bioeng Biotechnol. 2022 Jul 1;10:918659. doi: 10.3389/fbioe.2022.918659. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35845409 Free PMC article. Review.
References
-
- Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T. Nat. Biotechnol. 2001;19:751–755. - PubMed
-
- Forster A. C., Weissbach H., Blacklow S. C. Anal. Biochem. 2001;297:60–70. - PubMed
-
- Josephson K., Hartman M. C. T., Szostak J. W. J. Am. Chem. Soc. 2005;127:11727–11735. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

