Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1
- PMID: 16537382
- PMCID: PMC1450230
- DOI: 10.1073/pnas.0510403103
Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1
Erratum in
- Proc Natl Acad Sci U S A. 2006 Apr 25;103(17):6776
Abstract
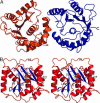
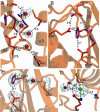
The ubiquitin C-terminal hydrolase UCH-L1 (PGP9.5) comprises >1% of total brain protein but is almost absent from other tissues [Wilkinson, K. D., et al. (1989) Science 246, 670-673]. Mutations in the UCH-L1 gene have been reported to be linked to susceptibility to and protection from Parkinson's disease [Leroy, E., et al. (1998) Nature 395, 451-452; Maraganore, D. M., et al. (1999) Neurology 53, 1858-1860]. Abnormal overexpression of UCH-L1 has been shown to correlate with several forms of cancer [Hibi, K., et al. (1998) Cancer Res. 58, 5690-5694]. Because the amino acid sequence of UCH-L1 is similar to that of other ubiquitin C-terminal hydrolases, including the ubiquitously expressed UCH-L3, which appear to be unconnected to neurodegenerative disease, the structure of UCH-L1 and the effects of disease associated mutations on the structure and function are of considerable importance. We have determined the three-dimensional structure of human UCH-L1 at 2.4-A resolution by x-ray crystallography. The overall fold resembles that of other ubiquitin hydrolases, including UCH-L3, but there are a number of significant differences. In particular, the geometry of the catalytic residues in the active site of UCH-L1 is distorted in such a way that the hydrolytic activity would appear to be impossible without substrate induced conformational rearrangements.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


Similar articles
-
Backbone and side-chain 1H, 15N and 13C resonance assignments of S18Y mutant of ubiquitin carboxy-terminal hydrolase L1.Biomol NMR Assign. 2011 Oct;5(2):165-8. doi: 10.1007/s12104-011-9292-7. Epub 2011 Feb 5. Biomol NMR Assign. 2011. PMID: 21298373 Free PMC article.
-
Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate.J Biol Chem. 2005 Jan 14;280(2):1512-20. doi: 10.1074/jbc.M410770200. Epub 2004 Nov 5. J Biol Chem. 2005. PMID: 15531586
-
Substrate recognition and catalysis by UCH-L1.Biochemistry. 2006 Dec 12;45(49):14717-25. doi: 10.1021/bi061406c. Biochemistry. 2006. PMID: 17144664
-
Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction.Biochem J. 2016 Aug 15;473(16):2453-62. doi: 10.1042/BCJ20160082. Biochem J. 2016. PMID: 27515257 Free PMC article. Review.
-
Familial Mutations and Post-translational Modifications of UCH-L1 in Parkinson's Disease and Neurodegenerative Disorders.Curr Protein Pept Sci. 2017;18(7):733-745. doi: 10.2174/1389203717666160217143721. Curr Protein Pept Sci. 2017. PMID: 26899237 Review.
Cited by
-
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV.ACS Chem Biol. 2015 Jun 19;10(6):1456-65. doi: 10.1021/cb500917m. Epub 2015 Mar 16. ACS Chem Biol. 2015. PMID: 25746232 Free PMC article.
-
Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3.J Biol Chem. 2010 Feb 26;285(9):6857-66. doi: 10.1074/jbc.M109.072405. Epub 2009 Dec 30. J Biol Chem. 2010. PMID: 20042598 Free PMC article.
-
Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation.Proc Natl Acad Sci U S A. 2010 May 18;107(20):9117-22. doi: 10.1073/pnas.0910870107. Epub 2010 May 3. Proc Natl Acad Sci U S A. 2010. PMID: 20439756 Free PMC article.
-
Gamma-glutamyl hydrolase: kinetic characterization of isopeptide hydrolysis using fluorogenic substrates.Biochemistry. 2008 Jan 29;47(4):1228-39. doi: 10.1021/bi701607v. Epub 2008 Jan 3. Biochemistry. 2008. PMID: 18171026 Free PMC article.
-
Loss of UCHL1 promotes age-related degenerative changes in the enteric nervous system.Front Aging Neurosci. 2014 Jun 19;6:129. doi: 10.3389/fnagi.2014.00129. eCollection 2014. Front Aging Neurosci. 2014. PMID: 24994982 Free PMC article.
References
-
- Hershko A., Ciechanover A., Varshavsky A. Nat. Med. 2000;6:1073–1081. - PubMed
-
- Amerik A. Y., Hochstrasser M. Biochim. Biophys. Acta. 2004;1695:189–207. - PubMed
-
- Chung C. H., Baek S. H. Biochem. Biophys. Res. Commun. 1999;266:633–640. - PubMed
-
- Wing S. S. Int. J. Biochem. Cell Biol. 2003;35:590–605. - PubMed
-
- Wilkinson K. D., Lee K. M., Deshpande S., Duerksen-Hughes P., Boss J. M., Pohl J. Science. 1989;246:670–673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

