Differential effects of heterochromatin protein 1 isoforms on mitotic chromosome distribution and growth in Dictyostelium discoideum
- PMID: 16524908
- PMCID: PMC1398066
- DOI: 10.1128/EC.5.3.530-543.2006
Differential effects of heterochromatin protein 1 isoforms on mitotic chromosome distribution and growth in Dictyostelium discoideum
Abstract
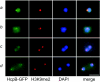
Heterochromatin protein 1 (HP1) is a well-characterized heterochromatin component conserved from fission yeast to humans. We identified three HP1-like genes (hcpA, hcpB, and hcpC) in the Dictyostelium discoideum genome. Two of these (hcpA and hcpB) are expressed, and the proteins colocalized as green fluorescent protein (GFP) fusion proteins in one major cluster at the nuclear periphery that was also characterized by histone H3 lysine 9 dimethylation, a histone modification so far not described for Dictyostelium. The data strongly suggest that this cluster represents the centromeres. Both single-knockout strains displayed only subtle phenotypes, suggesting that both isoforms have largely overlapping functions. In contrast, disruption of both isoforms appeared to be lethal. Furthermore, overexpression of a C-terminally truncated form of HcpA resulted in phenotypically distinct growth defects that were characterized by a strong decrease in cell viability. Although genetic evidence implies functional redundancy, overexpression of GFP-HcpA, but not GFP-HcpB, caused growth defects that were accompanied by an increase in the frequency of atypic anaphase bridges. Our data indicate that Dictyostelium discoideum cells are sensitive to changes in HcpA and HcpB protein levels and that the two isoforms display different in vivo and in vitro affinities for each other. Since the RNA interference (RNAi) machinery is frequently involved in chromatin remodeling, we analyzed if knockouts of RNAi components influenced the localization of H3K9 dimethylation and HP1 isoforms in Dictyostelium. Interestingly, heterochromatin organization appeared to be independent of functional RNAi.
Figures





Similar articles
-
Localization and organization of protein factors involved in chromosome inheritance in Dictyostelium discoideum.Biol Chem. 2007 Apr;388(4):355-65. doi: 10.1515/BC.2007.047. Biol Chem. 2007. PMID: 17391056
-
Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase.J Cell Sci. 2003 Aug 15;116(Pt 16):3327-38. doi: 10.1242/jcs.00635. Epub 2003 Jul 2. J Cell Sci. 2003. PMID: 12840071
-
Maintenance of stable heterochromatin domains by dynamic HP1 binding.Science. 2003 Jan 31;299(5607):721-5. doi: 10.1126/science.1078572. Science. 2003. PMID: 12560555
-
Heterochromatin protein 1: don't judge the book by its cover!Curr Opin Genet Dev. 2006 Apr;16(2):143-50. doi: 10.1016/j.gde.2006.02.013. Epub 2006 Feb 28. Curr Opin Genet Dev. 2006. PMID: 16503133 Review.
-
The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription.Bioessays. 2011 Apr;33(4):280-9. doi: 10.1002/bies.201000138. Epub 2011 Jan 27. Bioessays. 2011. PMID: 21271610 Review.
Cited by
-
Cell cycle regulated transcription of heterochromatin in mammals vs. fission yeast: functional conservation or coincidence?Cell Cycle. 2008 Jul 1;7(13):1907-10. doi: 10.4161/cc.7.13.6206. Epub 2008 Apr 29. Cell Cycle. 2008. PMID: 18604169 Free PMC article.
-
Convergent evolution of integration site selection upstream of tRNA genes by yeast and amoeba retrotransposons.Nucleic Acids Res. 2018 Aug 21;46(14):7250-7260. doi: 10.1093/nar/gky582. Nucleic Acids Res. 2018. PMID: 29945249 Free PMC article.
-
Structural and biochemical advances in mammalian RNAi.J Cell Biochem. 2006 Dec 1;99(5):1251-66. doi: 10.1002/jcb.21069. J Cell Biochem. 2006. PMID: 16927374 Free PMC article. Review.
-
Functional characterization of CP148, a novel key component for centrosome integrity in Dictyostelium.Cell Mol Life Sci. 2012 Jun;69(11):1875-88. doi: 10.1007/s00018-011-0904-2. Epub 2012 Jan 6. Cell Mol Life Sci. 2012. PMID: 22223109 Free PMC article.
-
Evolution of the nucleus.Curr Opin Cell Biol. 2014 Jun;28(100):8-15. doi: 10.1016/j.ceb.2014.01.004. Epub 2014 Feb 6. Curr Opin Cell Biol. 2014. PMID: 24508984 Free PMC article. Review.
References
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
-
- Bernard, P., J. F. Maure, J. F. Partridge, S. Genier, J. P. Javerzat, and R. C. Allshire. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539-2542. - PubMed
-
- Brasher, S. V., B. O. Smith, R. H. Fogh, D. Nietlispach, A. Thiru, P. R. Nielsen, R. W. Broadhurst, L. J. Ball, N. V. Murzina, and E. D. Laue. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19:1587-1597. - PMC - PubMed
-
- Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

