A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis
- PMID: 16516837
- PMCID: PMC1773005
- DOI: 10.1016/j.devcel.2006.01.012
A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis
Abstract
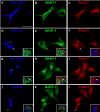
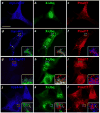
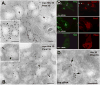
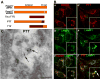
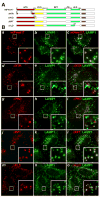
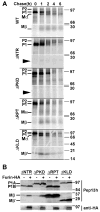
Cargo partitioning into intralumenal vesicles (ILVs) of multivesicular endosomes underlies such cellular processes as receptor downregulation, viral budding, and biogenesis of lysosome-related organelles such as melanosomes. We show that the melanosomal protein Pmel17 is sorted into ILVs by a mechanism that is dependent upon lumenal determinants and conserved in non-pigment cells. Pmel17 targeting to ILVs does not require its native cytoplasmic domain or cytoplasmic residues targeted by ubiquitylation and, unlike sorting of ubiquitylated cargo, is insensitive to functional inhibition of Hrs and ESCRT complexes. Chimeric protein and deletion analyses indicate that two N-terminal lumenal subdomains are necessary and sufficient for ILV targeting. Pmel17 fibril formation, which occurs during melanosome maturation in melanocytes, requires a third lumenal subdomain and proteolytic processing that itself requires ILV localization. These results establish an Hrs- and perhaps ESCRT-independent pathway of ILV sorting by lumenal determinants and a requirement for ILV sorting in fibril formation.
Figures
Similar articles
-
The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis.Dev Cell. 2011 Oct 18;21(4):708-21. doi: 10.1016/j.devcel.2011.08.019. Epub 2011 Sep 29. Dev Cell. 2011. PMID: 21962903 Free PMC article.
-
ESCRT-I function is required for Tyrp1 transport from early endosomes to the melanosome limiting membrane.Traffic. 2009 Sep;10(9):1318-36. doi: 10.1111/j.1600-0854.2009.00955.x. Epub 2009 Jun 9. Traffic. 2009. PMID: 19624486 Free PMC article.
-
Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells.J Cell Biol. 2001 Feb 19;152(4):809-24. doi: 10.1083/jcb.152.4.809. J Cell Biol. 2001. PMID: 11266471 Free PMC article.
-
ALIX and the multivesicular endosome: ALIX in Wonderland.Trends Cell Biol. 2014 Jan;24(1):19-25. doi: 10.1016/j.tcb.2013.10.009. Epub 2013 Nov 26. Trends Cell Biol. 2014. PMID: 24287454 Review.
-
Multivesicular bodies: co-ordinated progression to maturity.Curr Opin Cell Biol. 2008 Aug;20(4):408-14. doi: 10.1016/j.ceb.2008.04.001. Epub 2008 May 24. Curr Opin Cell Biol. 2008. PMID: 18502633 Free PMC article. Review.
Cited by
-
Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes.PLoS Biol. 2007 Jun;5(6):e158. doi: 10.1371/journal.pbio.0050158. PLoS Biol. 2007. PMID: 17550307 Free PMC article.
-
The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis.Proc Natl Acad Sci U S A. 2009 Aug 18;106(33):13731-6. doi: 10.1073/pnas.0906509106. Epub 2009 Jul 31. Proc Natl Acad Sci U S A. 2009. PMID: 19666488 Free PMC article.
-
Secretory granule membrane protein recycles through multivesicular bodies.Traffic. 2010 Jul 1;11(7):972-86. doi: 10.1111/j.1600-0854.2010.01066.x. Epub 2010 Apr 1. Traffic. 2010. PMID: 20374556 Free PMC article.
-
Transferrin-directed internalization and cycling of transferrin receptor 2.Traffic. 2009 Oct;10(10):1488-501. doi: 10.1111/j.1600-0854.2009.00961.x. Epub 2009 Jul 6. Traffic. 2009. PMID: 19682329 Free PMC article.
-
Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5.Mol Biol Cell. 2007 Feb;18(2):707-20. doi: 10.1091/mbc.e06-08-0680. Epub 2006 Dec 20. Mol Biol Cell. 2007. PMID: 17182849 Free PMC article.
References
-
- Arnaoutova I, Smith AM, Coates LC, Sharpe JC, Dhanvantari S, Snell CR, Birch NP, Loh YP. The prohormone processing enzyme PC3 is a lipid raft-associated transmembrane protein. Biochemistry. 2003;42:10445–10455. - PubMed
-
- Babst M. A protein’s final ESCRT. Traffic. 2005;6:2–9. - PubMed
-
- Bache KG, Raiborg C, Mehlum A, Stenmark H. STAM and Hrs are subunits of a multivalent Ubiquitin-binding complex on early endosomes. J Biol Chem. 2003b;278:12513–12521. - PubMed
-
- Berson JF, Frank DW, Calvo PA, Bieler BM, Marks MS. A common temperature-sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J Biol Chem. 2000;275:12281–12289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

