R5 and X4 HIV envelopes induce distinct gene expression profiles in primary peripheral blood mononuclear cells
- PMID: 16505369
- PMCID: PMC1533779
- DOI: 10.1073/pnas.0511237103
R5 and X4 HIV envelopes induce distinct gene expression profiles in primary peripheral blood mononuclear cells
Abstract
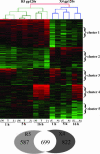
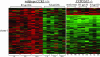
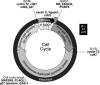
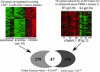
HIV envelope binds to and signals through its primary cellular receptor, CD4, and through a coreceptor, either CC chemokine receptor 5 (CCR5) or CXC chemokine receptor 4 (CXCR4). Here, we evaluate the response of peripheral blood mononuclear cells to a panel of genetically diverse R5 and X4 envelope proteins. Modulation of gene expression was evaluated by using oligonucleotide microarrays. Activation of transcription factors was evaluated by using an array of oligonucleotides encoding transcription factor binding sites. Responses were strongly influenced by coreceptor specificity. Treatment of cells from CCR5delta32 homozygous donors with glycoprotein (gp)120 derived from an R5 virus demonstrated that the majority of responses elicited by R5 envelopes required engagement of CCR5. R5 envelopes, to a greater extent than X4 envelopes, induced the expression of genes belonging to mitogen-activated protein kinase signal transduction pathways and genes regulating the cell cycle. A number of genes induced by R5, but not X4, envelopes were also up-regulated in the resting CD4+ T cell population of HIV-infected individuals. These results suggest that R5 envelope facilitates replication of HIV in the pool of resting CD4+ T cells. Additionally, signaling by R5 gp120 may facilitate the transmission of R5 viruses by inducing a permissive environment for HIV replication.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


Similar articles
-
R5 HIV gp120-mediated cellular contacts induce the death of single CCR5-expressing CD4 T cells by a gp41-dependent mechanism.J Leukoc Biol. 2004 Oct;76(4):804-11. doi: 10.1189/jlb.0204100. Epub 2004 Jul 16. J Leukoc Biol. 2004. PMID: 15258189
-
Differential effects of R5 and X4 human immunodeficiency virus type 1 infection on CD4+ cell proliferation and activation.J Gen Virol. 2005 Apr;86(Pt 4):1171-1179. doi: 10.1099/vir.0.80674-0. J Gen Virol. 2005. PMID: 15784911
-
Envelope-dependent restriction of human immunodeficiency virus type 1 spreading in CD4(+) T lymphocytes: R5 but not X4 viruses replicate in the absence of T-cell receptor restimulation.J Virol. 1999 Sep;73(9):7515-23. doi: 10.1128/JVI.73.9.7515-7523.1999. J Virol. 1999. PMID: 10438841 Free PMC article.
-
Relationships Between HIV-Mediated Chemokine Coreceptor Signaling, Cofilin Hyperactivation, Viral Tropism Switch and HIV-Mediated CD4 Depletion.Curr HIV Res. 2019;17(6):388-396. doi: 10.2174/1570162X17666191106112018. Curr HIV Res. 2019. PMID: 31702526 Review.
-
HIV-1 infection and chemokine receptor modulation.Curr HIV Res. 2004 Jan;2(1):39-50. doi: 10.2174/1570162043484997. Curr HIV Res. 2004. PMID: 15053339 Review.
Cited by
-
Asymmetric HIV-1 co-receptor use and replication in CD4(+) T lymphocytes.J Transl Med. 2011 Jan 27;9 Suppl 1(Suppl 1):S8. doi: 10.1186/1479-5876-9-S1-S8. J Transl Med. 2011. PMID: 21284907 Free PMC article. Review.
-
HIV envelope gp120 activates LFA-1 on CD4 T-lymphocytes and increases cell susceptibility to LFA-1-targeting leukotoxin (LtxA).PLoS One. 2011;6(8):e23202. doi: 10.1371/journal.pone.0023202. Epub 2011 Aug 5. PLoS One. 2011. PMID: 21850260 Free PMC article.
-
CCR5Delta32 protein expression and stability are critical for resistance to human immunodeficiency virus type 1 in vivo.J Virol. 2007 Aug;81(15):8041-9. doi: 10.1128/JVI.00068-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522201 Free PMC article.
-
The HIV-1 gp120/V3 modifies the response of uninfected CD4 T cells to antigen presentation: mapping of the specific transcriptional signature.J Transl Med. 2011 Sep 24;9:160. doi: 10.1186/1479-5876-9-160. J Transl Med. 2011. PMID: 21943198 Free PMC article.
-
Unfolding the relationship between secreted molecular chaperones and macrophage activation states.Cell Stress Chaperones. 2009 Jul;14(4):329-41. doi: 10.1007/s12192-008-0087-4. Epub 2008 Oct 29. Cell Stress Chaperones. 2009. PMID: 18958583 Free PMC article. Review.
References
-
- Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., Di Marzio P., Marmon S., Sutton R. E., Hill C. M., et al. Nature. 1996;381:661–666. - PubMed
-
- Dragic T., Litwin V., Allaway G. P., Martin S. R., Huang Y., Nagashima K. A., Cayanan C., Maddon P. J., Koup R. A., Moore J. P., Paxton W. A. Nature. 1996;381:667–673. - PubMed
-
- Feng Y., Broder C. C., Kennedy P. E., Berger E. A. Science. 1996;272:872–877. - PubMed
-
- Juszczak R. J., Turchin H., Truneh A., Culp J., Kassis S. J. Biol. Chem. 1991;266:11176–11183. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

