Identification of mouse orthologue of endogenous secretory receptor for advanced glycation end-products: structure, function and expression
- PMID: 16503878
- PMCID: PMC1450004
- DOI: 10.1042/BJ20051573
Identification of mouse orthologue of endogenous secretory receptor for advanced glycation end-products: structure, function and expression
Abstract
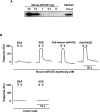
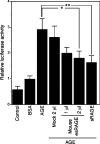
The cell-surface RAGE [receptor for AGE (advanced glycation end-products)] is associated with the development of diabetic vascular complications, neurodegenerative disorders and inflammation. Recently, we isolated a human RAGE splice variant, which can work as a decoy receptor for RAGE ligands, and named it esRAGE (endogenous secretory RAGE). In the present study, we have isolated the murine equivalent of esRAGE from brain polysomal poly(A)+ (polyadenylated) RNA by RT (reverse transcription)-PCR cloning. The mRNA was generated by alternative splicing, and it encoded a 334-amino-acid protein with a signal sequence, but lacking the transmembrane domain. A transfection experiment revealed that the mRNA was actually translated as deduced to yield the secretory protein working as a decoy in AGE-induced NF-kappaB (nuclear factor kappaB) activation. RT-PCR and immunoblotting detected esRAGE mRNA and protein in the brain, lung, kidney and small intestine of wild-type mice, but not of RAGE-null mice. The esRAGE expression was increased in the kidney of diabetic wild-type mice. The present study has thus provided an animal orthologue of esRAGE for clarification of its roles in health and disease.
Figures



Similar articles
-
Novel splice variants of the receptor for advanced glycation end-products expressed in human vascular endothelial cells and pericytes, and their putative roles in diabetes-induced vascular injury.Biochem J. 2003 Mar 15;370(Pt 3):1097-109. doi: 10.1042/BJ20021371. Biochem J. 2003. PMID: 12495433 Free PMC article.
-
Receptor for advanced glycation end products is involved in impaired angiogenic response in diabetes.Diabetes. 2006 Aug;55(8):2245-55. doi: 10.2337/db05-1375. Diabetes. 2006. PMID: 16873687
-
Endogenous secretory receptor for advanced glycation end-products and cardiovascular disease in end-stage renal disease.J Ren Nutr. 2008 Jan;18(1):76-82. doi: 10.1053/j.jrn.2007.10.016. J Ren Nutr. 2008. PMID: 18089449 Review.
-
Receptor for advanced glycation end products (RAGE) mediates neuronal differentiation and neurite outgrowth.J Neurosci Res. 2008 May 1;86(6):1254-66. doi: 10.1002/jnr.21578. J Neurosci Res. 2008. PMID: 18058943
-
RAGE axis: Animal models and novel insights into the vascular complications of diabetes.Arterioscler Thromb Vasc Biol. 2004 Aug;24(8):1342-9. doi: 10.1161/01.ATV.0000133191.71196.90. Epub 2004 May 20. Arterioscler Thromb Vasc Biol. 2004. PMID: 15155381 Review.
Cited by
-
The Mouse-Specific Splice Variant mRAGE_v4 Encodes a Membrane-Bound RAGE That Is Resistant to Shedding and Does Not Contribute to the Production of Soluble RAGE.PLoS One. 2016 Sep 21;11(9):e0153832. doi: 10.1371/journal.pone.0153832. eCollection 2016. PLoS One. 2016. PMID: 27655137 Free PMC article.
-
High-mobility group box-1 protein and β-amyloid oligomers promote neuronal differentiation of adult hippocampal neural progenitors via receptor for advanced glycation end products/nuclear factor-κB axis: relevance for Alzheimer's disease.J Neurosci. 2013 Apr 3;33(14):6047-59. doi: 10.1523/JNEUROSCI.2052-12.2013. J Neurosci. 2013. PMID: 23554486 Free PMC article.
-
Soluble receptor for advanced glycation end products protects from ischemia- and reperfusion-induced acute kidney injury.Biol Open. 2022 Jan 15;11(1):bio058852. doi: 10.1242/bio.058852. Epub 2022 Feb 4. Biol Open. 2022. PMID: 34812852 Free PMC article.
-
Blockade of RAGE ameliorates elastase-induced emphysema development and progression via RAGE-DAMP signaling.FASEB J. 2017 May;31(5):2076-2089. doi: 10.1096/fj.201601155R. Epub 2017 Feb 1. FASEB J. 2017. PMID: 28148566 Free PMC article.
-
Modulation of RAGE isoforms expression in the brain and plasma of rats exposed to transient focal cerebral ischemia.Neurochem Res. 2012 Jul;37(7):1508-16. doi: 10.1007/s11064-012-0778-1. Epub 2012 Apr 19. Neurochem Res. 2012. PMID: 22528836
References
-
- Schmidt A. M., Stern D. M. RAGE: a new target for the prevention and treatment of the vascular and inflammatory complications of diabetes. Trends Endocrinol. Metab. 2000;11:368–375. - PubMed
-
- Neeper M., Schmidt A. M., Brett J., Yan S. D., Wang F., Pan Y. C. E., Elliston K., Stern D., Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J. Biol. Chem. 1992;267:14998–15004. - PubMed
-
- Hofmann M. A., Drury S., Fu C., Qu W., Taguchi A., Lu Y., Avil C., Kambham N., Bierhaus A., Nawroth P., et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. - PubMed
-
- Huttunen H. J., Fages C., Rauvala H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-κB required the cytoplasmic domain of the receptor but different downstream signaling pathway. J. Biol. Chem. 1999;274:19919–19924. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

