The geometry of DNA supercoils modulates topoisomerase-mediated DNA cleavage and enzyme response to anticancer drugs
- PMID: 16503659
- PMCID: PMC2517258
- DOI: 10.1021/bi051987q
The geometry of DNA supercoils modulates topoisomerase-mediated DNA cleavage and enzyme response to anticancer drugs
Abstract
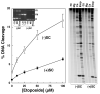
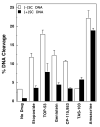
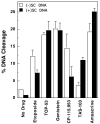
Collisions with DNA tracking systems are critical for the conversion of transient topoisomerase-DNA cleavage complexes to permanent strand breaks. Since DNA is overwound ahead of tracking systems, cleavage complexes most likely to produce permanent strand breaks should be formed between topoisomerases and positively supercoiled molecules. Therefore, the ability of human topoisomerase IIalpha and IIbeta and topoisomerase I to cleave positively supercoiled DNA was assessed in the absence or presence of anticancer drugs. Topoisomerase IIalpha and IIbeta maintained approximately 4-fold lower levels of cleavage complexes with positively rather than negatively supercoiled DNA. Topoisomerase IIalpha also displayed lower levels of cleavage with overwound substrates in the presence of nonintercalative drugs. Decreased drug efficacy was due primarily to a drop in baseline (i.e., nondrug) cleavage, rather than an altered interaction with the enzyme-DNA complex. Similar results were seen for topoisomerase IIbeta, but the effects of DNA geometry on drug-induced scission were somewhat less pronounced. With both topoisomerase IIalpha and IIbeta, intercalative drugs displayed greater relative cleavage enhancement with positively supercoiled DNA. This appeared to result from negative effects of high concentrations of intercalative agents on underwound DNA. In contrast to the type II enzymes, topoisomerase I maintained approximately 3-fold higher levels of cleavage complexes with positively supercoiled substrates and displayed an even more dramatic increase in the presence of camptothecin. These findings suggest that the geometry of DNA supercoils has a profound influence on topoisomerase-mediated DNA scission and that topoisomerase I may be an intrinsically more lethal target for anticancer drugs than either topoisomerase IIalpha or IIbeta.
Figures





Similar articles
-
Bimodal recognition of DNA geometry by human topoisomerase II alpha: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain.Biochemistry. 2008 Dec 16;47(50):13169-78. doi: 10.1021/bi800453h. Biochemistry. 2008. PMID: 19053267 Free PMC article.
-
The geometry of DNA supercoils modulates the DNA cleavage activity of human topoisomerase I.Nucleic Acids Res. 2011 Feb;39(3):1014-22. doi: 10.1093/nar/gkq822. Epub 2010 Sep 19. Nucleic Acids Res. 2011. PMID: 20855291 Free PMC article.
-
Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks.J Biol Chem. 2005 Nov 25;280(47):39337-45. doi: 10.1074/jbc.M503320200. Epub 2005 Sep 27. J Biol Chem. 2005. PMID: 16188892
-
Oligonucleotide-Recognizing Topoisomerase Inhibitors (OTIs): Precision Gene Editors for Neurodegenerative Diseases?Int J Mol Sci. 2022 Sep 29;23(19):11541. doi: 10.3390/ijms231911541. Int J Mol Sci. 2022. PMID: 36232843 Free PMC article. Review.
-
Telling Your Right Hand from Your Left: The Effects of DNA Supercoil Handedness on the Actions of Type II Topoisomerases.Int J Mol Sci. 2023 Jul 7;24(13):11199. doi: 10.3390/ijms241311199. Int J Mol Sci. 2023. PMID: 37446377 Free PMC article. Review.
Cited by
-
Bimodal recognition of DNA geometry by human topoisomerase II alpha: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain.Biochemistry. 2008 Dec 16;47(50):13169-78. doi: 10.1021/bi800453h. Biochemistry. 2008. PMID: 19053267 Free PMC article.
-
Interactions between the etoposide derivative F14512 and human type II topoisomerases: implications for the C4 spermine moiety in promoting enzyme-mediated DNA cleavage.Biochemistry. 2011 Apr 19;50(15):3240-9. doi: 10.1021/bi200094z. Epub 2011 Mar 28. Biochemistry. 2011. PMID: 21413765 Free PMC article.
-
Amsacrine as a topoisomerase II poison: importance of drug-DNA interactions.Biochemistry. 2012 Feb 28;51(8):1730-9. doi: 10.1021/bi201159b. Epub 2012 Feb 10. Biochemistry. 2012. PMID: 22304499 Free PMC article.
-
DNA cleavage assay for the identification of topoisomerase I inhibitors.Nat Protoc. 2008;3(11):1736-50. doi: 10.1038/nprot.2008.174. Nat Protoc. 2008. PMID: 18927559 Free PMC article.
-
DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition.Chem Rev. 2009 Jul;109(7):2894-902. doi: 10.1021/cr900097c. Chem Rev. 2009. PMID: 19476377 Free PMC article. Review. No abstract available.
References
-
- Osheroff N. DNA topoisomerases. Biochim. Biophys. Acta. 1998;1400:1–2. - PubMed
-
- Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta. 1998;1400:63–81. - PubMed
-
- Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. - PubMed
-
- Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:221–253. - PubMed
-
- Champoux JJ. DNA topisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

