A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling
- PMID: 16497831
- PMCID: PMC1533784
- DOI: 10.1073/pnas.0600007103
A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling
Abstract
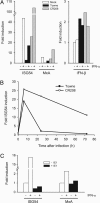
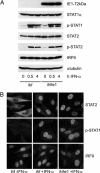
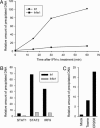
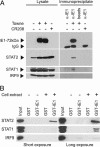
Type I IFNs are crucial components of the innate immune response to viral attack. They are rapidly synthesized and secreted after infection with human cytomegalovirus (CMV) and trigger a signal transduction pathway that involves successive activation and nuclear translocation of signal transducer and activator of transcription 1 (STAT1) and STAT2. The activated STATs, together with the IFN regulatory factor 9 protein, form a trimeric transcription complex (IFN-stimulated gene factor 3) that stimulates expression of numerous IFN-responsive genes, many of which exhibit antiviral activity. Here we demonstrate that the viral 72-kDa IE1 protein (IE1-72kDa) confers partial resistance to the antiviral activity of type I IFNs upon CMV. Accordingly, IFN-responsive transcripts accumulate to substantially increased levels after infection with an IE1-deficient mutant as compared with wild-type virus, and ectopic expression of the viral protein in stably transfected cells is sufficient to block their induction. We further show that IE1-72kDa forms a physical complex with STAT1 and STAT2 in nuclei of infected cells and in vitro and prevents association of STAT1, STAT2, and IFN regulatory factor 9 with promoters of IFN-responsive genes in vivo. Our results indicate that the viral protein blocks an intranuclear step after nuclear translocation and before DNA binding of IFN-stimulated gene factor 3, presumably by interfering with the integrity and/or correct subnuclear localization of the protein complex. This study identifies the CMV IE1-72kDa protein as a viral antagonist of the cellular innate immune response, inhibiting IFN-dependent STAT signaling by means of an unprecedented molecular mechanism.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


Similar articles
-
The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2.Virology. 2001 May 10;283(2):230-9. doi: 10.1006/viro.2001.0856. Virology. 2001. PMID: 11336548
-
IFN-type-I-mediated signaling is regulated by modulation of STAT2 nuclear export.J Cell Sci. 2006 Mar 15;119(Pt 6):1092-104. doi: 10.1242/jcs.02822. Epub 2006 Feb 28. J Cell Sci. 2006. PMID: 16507591
-
IRF-9/STAT2 [corrected] functional interaction drives retinoic acid-induced gene G expression independently of STAT1.Cancer Res. 2009 Apr 15;69(8):3673-80. doi: 10.1158/0008-5472.CAN-08-4922. Epub 2009 Apr 7. Cancer Res. 2009. PMID: 19351818
-
The interferon receptors.Semin Oncol. 1997 Jun;24(3 Suppl 9):S9-18-S9-40. Semin Oncol. 1997. PMID: 9208871 Review.
-
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
Cited by
-
The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction.Front Cell Dev Biol. 2022 Nov 25;10:1053139. doi: 10.3389/fcell.2022.1053139. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506089 Free PMC article. Review.
-
HCMV-encoded US7 and US8 act as antagonists of innate immunity by distinctively targeting TLR-signaling pathways.Nat Commun. 2019 Oct 11;10(1):4670. doi: 10.1038/s41467-019-12641-4. Nat Commun. 2019. PMID: 31604943 Free PMC article.
-
Transmembrane Protein pUL50 of Human Cytomegalovirus Inhibits ISGylation by Downregulating UBE1L.J Virol. 2018 Jul 17;92(15):e00462-18. doi: 10.1128/JVI.00462-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29743376 Free PMC article.
-
Foot-and-mouth disease virus structural protein VP3 degrades Janus kinase 1 to inhibit IFN-γ signal transduction pathways.Cell Cycle. 2016;15(6):850-60. doi: 10.1080/15384101.2016.1151584. Cell Cycle. 2016. PMID: 26901336 Free PMC article.
-
Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection.J Virol. 2008 Jan;82(1):126-37. doi: 10.1128/JVI.01685-07. Epub 2007 Oct 17. J Virol. 2008. PMID: 17942542 Free PMC article.
References
-
- Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. Annu. Rev. Biochem. 1998;67:227–264. - PubMed
-
- Weber F., Kochs G., Haller O. Viral Immunol. 2004;17:498–515. - PubMed
-
- Biron C. A., Sen G. C. In: Fields Virology. Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E., editors. Vol. 1. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 321–351.
-
- Rawlings J. S., Rosler K. M., Harrison D. A. J. Cell Sci. 2004;117:1281–1283. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

