Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen
- PMID: 16492767
- PMCID: PMC1413915
- DOI: 10.1073/pnas.0511224103
Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen
Abstract
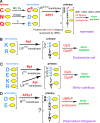
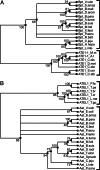
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. Primary destabilizing N-terminal residues (Nd(p)) are recognized directly by the targeting machinery. The recognition of secondary destabilizing N-terminal residues (Nd(s)) is preceded by conjugation of an Nd(p) residue to Nd(s) of a polypeptide substrate. In eukaryotes, ATE1-encoded arginyl-transferases (R(D,E,C*)-transferases) conjugate Arg (R), an Nd(p) residue, to Nd(s) residues Asp (D), Glu (E), or oxidized Cys residue (C*). Ubiquitin ligases recognize the N-terminal Arg of a substrate and target the (ubiquitylated) substrate to the proteasome. In prokaryotes such as Escherichia coli, Nd(p) residues Leu (L) or Phe (F) are conjugated, by the aat-encoded Leu/Phe-transferase (L/F(K,R)-transferase), to N-terminal Arg or Lys, which are Nd(s) in prokaryotes but Nd(p) in eukaryotes. In prokaryotes, substrates bearing the Nd(p) residues Leu, Phe, Trp, or Tyr are degraded by the proteasome-like ClpAP protease. Despite enzymological similarities between eukaryotic R(D,E,C*)-transferases and prokaryotic L/F(K,R)-transferases, there is no significant sequelogy (sequence similarity) between them. We identified an aminoacyl-transferase, termed Bpt, in the human pathogen Vibrio vulnificus. Although it is a sequelog of eukaryotic R(D,E,C*)-transferases, this prokaryotic transferase exhibits a "hybrid" specificity, conjugating Nd(p) Leu to Nd(s) Asp or Glu. Another aminoacyl-transferase, termed ATEL1, of the eukaryotic pathogen Plasmodium falciparum, is a sequelog of prokaryotic L/F(K,R)-transferases (Aat), but has the specificity of eukaryotic R(D,E,C*)-transferases (ATE1). Phylogenetic analysis suggests that the substrate specificity of R-transferases arose by two distinct routes during the evolution of eukaryotes.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


Similar articles
-
Analyzing N-terminal Arginylation through the Use of Peptide Arrays and Degradation Assays.J Biol Chem. 2016 Sep 30;291(40):20976-20992. doi: 10.1074/jbc.M116.747956. Epub 2016 Aug 10. J Biol Chem. 2016. PMID: 27510035 Free PMC article.
-
Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway.Mol Cell Biol. 1999 Jan;19(1):182-93. doi: 10.1128/MCB.19.1.182. Mol Cell Biol. 1999. PMID: 9858543 Free PMC article.
-
Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog.EMBO J. 2006 Dec 13;25(24):5942-50. doi: 10.1038/sj.emboj.7601433. Epub 2006 Nov 16. EMBO J. 2006. PMID: 17110926 Free PMC article.
-
The molecular basis for the post-translational addition of amino acids by L/F transferase in the N-end rule pathway.Curr Protein Pept Sci. 2015;16(2):163-80. Curr Protein Pept Sci. 2015. PMID: 25692952 Review.
-
The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies.Trends Cell Biol. 2007 Apr;17(4):165-72. doi: 10.1016/j.tcb.2007.02.001. Epub 2007 Feb 15. Trends Cell Biol. 2007. PMID: 17306546 Review.
Cited by
-
Applications of Bacterial Degrons and Degraders - Toward Targeted Protein Degradation in Bacteria.Front Mol Biosci. 2021 May 7;8:669762. doi: 10.3389/fmolb.2021.669762. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026843 Free PMC article. Review.
-
Sequence and structure evolved separately in a ribosomal ubiquitin variant.EMBO J. 2007 Jul 25;26(14):3474-83. doi: 10.1038/sj.emboj.7601772. Epub 2007 Jun 28. EMBO J. 2007. PMID: 17599068 Free PMC article.
-
Identification of protein stability determinants in chloroplasts.Plant J. 2010 Aug;63(4):636-50. doi: 10.1111/j.1365-313X.2010.04268.x. Plant J. 2010. PMID: 20545891 Free PMC article.
-
The N-end rule pathway: emerging functions and molecular principles of substrate recognition.Nat Rev Mol Cell Biol. 2011 Oct 21;12(11):735-47. doi: 10.1038/nrm3217. Nat Rev Mol Cell Biol. 2011. PMID: 22016057 Review.
-
Analyzing N-terminal Arginylation through the Use of Peptide Arrays and Degradation Assays.J Biol Chem. 2016 Sep 30;291(40):20976-20992. doi: 10.1074/jbc.M116.747956. Epub 2016 Aug 10. J Biol Chem. 2016. PMID: 27510035 Free PMC article.
References
-
- Varshavsky A. Trends Biochem. Sci. 2005;30:283–286. - PubMed
-
- Hershko A., Ciechanover A., Varshavsky A. Nat. Med. 2000;10:1073–1081. - PubMed
-
- Pickart C. Cell. 2004;116:181–190. - PubMed
-
- Petroski M. D., Deshaies R. J. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. - PubMed
-
- Baumeister W., Walz J., Zühl F., Seemüller E. Cell. 1998;92:367–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

