The cytoplasmic C-terminal domain of the Escherichia coli KdpD protein functions as a K+ sensor
- PMID: 16484207
- PMCID: PMC1426542
- DOI: 10.1128/JB.188.5.1950-1958.2006
The cytoplasmic C-terminal domain of the Escherichia coli KdpD protein functions as a K+ sensor
Abstract
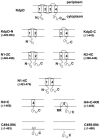
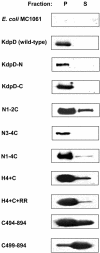
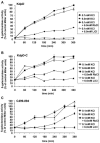
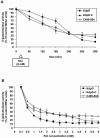
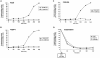
The KdpD protein is a K(+) sensor kinase located in the cytoplasmic membrane of Escherichia coli. It contains four transmembrane stretches and two short periplasmic loops of 4 and 10 amino acid residues, respectively. To determine which part of KdpD functions as a K(+) sensor, genetic variants were constructed with truncations or altered arrangements of the transmembrane segments. All KdpD constructs were tested by complementation of an E. coli kdpD deletion strain for their ability to grow at a K(+) concentration of 0.1 mM in the medium. A soluble protein composed of the C-terminal cytoplasmic domain was able to complement the kdpD deletion strain. In addition, analysis of the beta-galactosidase activity of an E. coli strain which carries a transcriptional fusion of the upstream region of the kdpFABC operon and a promoterless lacZ gene revealed that this soluble KdpD mutant responds to changes in the K(+) concentration in the extracellular medium. The results suggest that the sensing and response functions are both located in the C-terminal domain and might be modulated by the N-terminal domain as well as by membrane anchoring.
Figures
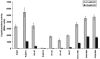
Similar articles
-
An atypical KdpD homologue from the cyanobacterium Anabaena sp. strain L-31: cloning, in vivo expression, and interaction with Escherichia coli KdpD-CTD.J Bacteriol. 2005 Jul;187(14):4921-7. doi: 10.1128/JB.187.14.4921-4927.2005. J Bacteriol. 2005. PMID: 15995207 Free PMC article.
-
The extension of the fourth transmembrane helix of the sensor kinase KdpD of Escherichia coli is involved in sensing.J Bacteriol. 2007 Oct;189(20):7326-34. doi: 10.1128/JB.00976-07. Epub 2007 Aug 17. J Bacteriol. 2007. PMID: 17704218 Free PMC article.
-
Amino acid replacements in transmembrane domain 1 influence osmosensing but not K+ sensing by the sensor kinase KdpD of Escherichia coli.Arch Microbiol. 2002 Dec;178(6):525-30. doi: 10.1007/s00203-002-0485-4. Epub 2002 Oct 3. Arch Microbiol. 2002. PMID: 12420175
-
The complexity of the 'simple' two-component system KdpD/KdpE in Escherichia coli.FEMS Microbiol Lett. 2010 Mar;304(2):97-106. doi: 10.1111/j.1574-6968.2010.01906.x. Epub 2010 Jan 20. FEMS Microbiol Lett. 2010. PMID: 20146748 Review.
-
Towards an understanding of the molecular mechanisms of stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli.J Mol Microbiol Biotechnol. 2002 May;4(3):223-8. J Mol Microbiol Biotechnol. 2002. PMID: 11931551 Review.
Cited by
-
Structure and function of the juxtamembrane GAF domain of potassium biosensor KdpD.Protein Sci. 2020 Sep;29(9):2009-2021. doi: 10.1002/pro.3920. Epub 2020 Aug 17. Protein Sci. 2020. PMID: 32713093 Free PMC article.
-
The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm.EMBO J. 2012 May 30;31(11):2648-59. doi: 10.1038/emboj.2012.99. Epub 2012 Apr 27. EMBO J. 2012. PMID: 22543870 Free PMC article.
-
SecA drives transmembrane insertion of RodZ, an unusual single-span membrane protein.J Mol Biol. 2015 Mar 13;427(5):1023-37. doi: 10.1016/j.jmb.2014.05.005. Epub 2014 May 15. J Mol Biol. 2015. PMID: 24846669 Free PMC article.
-
The Global Reciprocal Reprogramming between Mycobacteriophage SWU1 and Mycobacterium Reveals the Molecular Strategy of Subversion and Promotion of Phage Infection.Front Microbiol. 2016 Jan 28;7:41. doi: 10.3389/fmicb.2016.00041. eCollection 2016. Front Microbiol. 2016. PMID: 26858712 Free PMC article.
-
An amphiphilic region in the cytoplasmic domain of KdpD is recognized by the signal recognition particle and targeted to the Escherichia coli membrane.Mol Microbiol. 2008 Jun;68(6):1471-84. doi: 10.1111/j.1365-2958.2008.06246.x. Epub 2008 Apr 21. Mol Microbiol. 2008. PMID: 18433452 Free PMC article.
References
-
- Altendorf, K., and W. Epstein. 1996. The Kdp-ATPase of Escherichia coli, p. 403-420. In A. G. Lee (ed.), Biomembranes, vol. 5. Jai Press Ltd., London, England.
-
- Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. - PubMed
-
- Davis, N. G., and P. Model. 1985. An artificial anchor domain: hydrophobicity suffices to stop transfer. Cell 41:607-614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

