Phage display in the study of infectious diseases
- PMID: 16460941
- PMCID: PMC7127285
- DOI: 10.1016/j.tim.2006.01.006
Phage display in the study of infectious diseases
Abstract
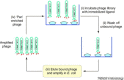
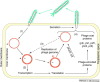
Microbial infections are dependent on the panoply of interactions between pathogen and host and identifying the molecular basis of such interactions is necessary to understand and control infection. Phage display is a simple functional genomic methodology for screening and identifying protein-ligand interactions and is widely used in epitope mapping, antibody engineering and screening for receptor agonists or antagonists. Phage display is also used widely in various forms, including the use of fragment libraries of whole microbial genomes, to identify peptide-ligand and protein-ligand interactions that are of importance in infection. In particular, this technique has proved successful in identifying microbial adhesins that are vital for colonization.
Figures

Similar articles
-
Epitope mapping by phage display: random versus gene-fragment libraries.J Immunol Methods. 1997 Aug 7;206(1-2):43-52. doi: 10.1016/s0022-1759(97)00083-5. J Immunol Methods. 1997. PMID: 9328567
-
Mapping of linear epitopes recognized by monoclonal antibodies with gene-fragment phage display libraries.Mol Gen Genet. 1995 Dec 10;249(4):425-31. doi: 10.1007/BF00287104. Mol Gen Genet. 1995. PMID: 8552047
-
Epitope mapping of Mycoplasma hyopneumoniae using phage displayed peptide libraries and the immune responses of the selected phagotopes.J Immunol Methods. 2005 Sep;304(1-2):15-29. doi: 10.1016/j.jim.2005.05.009. J Immunol Methods. 2005. PMID: 16054642
-
Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics.Curr Drug Targets. 2004 Jan;5(1):1-15. doi: 10.2174/1389450043490668. Curr Drug Targets. 2004. PMID: 14738215 Review.
-
Phage display for epitope determination: a paradigm for identifying receptor-ligand interactions.Biotechnol Annu Rev. 2004;10:151-88. doi: 10.1016/S1387-2656(04)10006-9. Biotechnol Annu Rev. 2004. PMID: 15504706 Review.
Cited by
-
The Chlamydia outer membrane protein OmcB is required for adhesion and exhibits biovar-specific differences in glycosaminoglycan binding.Mol Microbiol. 2008 Jan;67(2):403-19. doi: 10.1111/j.1365-2958.2007.06050.x. Epub 2007 Dec 11. Mol Microbiol. 2008. PMID: 18086188 Free PMC article.
-
Phage Display as a Medium for Target Therapy Based Drug Discovery, Review and Update.Mol Biotechnol. 2024 Jun 1. doi: 10.1007/s12033-024-01195-6. Online ahead of print. Mol Biotechnol. 2024. PMID: 38822912 Review.
-
Benchmarking bioinformatic virus identification tools using real-world metagenomic data across biomes.Genome Biol. 2024 Apr 15;25(1):97. doi: 10.1186/s13059-024-03236-4. Genome Biol. 2024. PMID: 38622738 Free PMC article.
-
Direct selection and phage display of a Gram-positive secretome.Genome Biol. 2007;8(12):R266. doi: 10.1186/gb-2007-8-12-r266. Genome Biol. 2007. PMID: 18078523 Free PMC article.
-
Triosephosphate isomerase of Taenia solium (TTPI): phage display and antibodies as tools for finding target regions to inhibit catalytic activity.Parasitol Res. 2015 Jan;114(1):55-64. doi: 10.1007/s00436-014-4159-3. Epub 2014 Oct 3. Parasitol Res. 2015. PMID: 25273631
References
-
- Smith G.P. Filamentous fusion phage – novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. - PubMed
-
- Russel M. Introduction to phage display and phage biology. In: Clackson T., Lowman H., editors. Phage Display: A Practical Approach. Oxford University Press; 2004. pp. 1–26.
-
- Azzazy H.M.E., Highsmith J.W.E. Phage display technology: clinical applications and recent innovations. Clin. Biochem. 2002;35:425–445. - PubMed
-
- Castagnoli L. Alternative bacteriophage display systems. Comb. Chem. High Throughput Screen. 2001;4:121–133. - PubMed
-
- Lindqvist B.H., Naderi S. Peptide presentation by bacteriophage P4. FEMS Microbiol. Rev. 1995;17:33–39. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

