RNA editing and alternative splicing: the importance of co-transcriptional coordination
- PMID: 16440002
- PMCID: PMC1456888
- DOI: 10.1038/sj.embor.7400621
RNA editing and alternative splicing: the importance of co-transcriptional coordination
Abstract
The carboxy-terminal domain (CTD) of the large subunit of RNA polymerase II (pol II) is essential for several co-transcriptional pre-messenger RNA processing events, including capping, 3'-end processing and splicing. We investigated the role of the CTD of RNA pol II in the coordination of A to I editing and splicing of the ADAR2 (ADAR: adenosine deaminases that act on RNA) pre-mRNA. The auto-editing of Adar2 intron 4 by the ADAR2 adenosine deaminase is tightly coupled to splicing, as the modification of the dinucleotide AA to AI creates a new 3' splice site. Unlike other introns, the CTD is not required for efficient splicing of intron 4 at either the normal 3' splice site or the alternative site created by editing. However, the CTD is required for efficient co-transcriptional auto-editing of ADAR2 intron 4. Our results implicate the CTD in site-selective RNA editing by ADAR2 and in coordination of editing with alternative splicing.
Figures

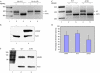

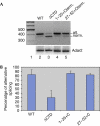
Comment in
-
An editor controlled by transcription.EMBO Rep. 2006 Mar;7(3):269-70. doi: 10.1038/sj.embor.7400650. EMBO Rep. 2006. PMID: 16607395 Free PMC article. No abstract available.
Similar articles
-
The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript.RNA. 2007 Jul;13(7):1071-8. doi: 10.1261/rna.404407. Epub 2007 May 24. RNA. 2007. PMID: 17525170 Free PMC article.
-
Structure and sequence determinants required for the RNA editing of ADAR2 substrates.J Biol Chem. 2004 Feb 6;279(6):4941-51. doi: 10.1074/jbc.M310068200. Epub 2003 Nov 30. J Biol Chem. 2004. PMID: 14660658
-
Phylogenetic comparison of the pre-mRNA adenosine deaminase ADAR2 genes and transcripts: conservation and diversity in editing site sequence and alternative splicing patterns.Gene. 2002 Oct 16;299(1-2):83-94. doi: 10.1016/s0378-1119(02)01016-8. Gene. 2002. PMID: 12459255
-
ADAR RNA editing in innate immune response phasing, in circadian clocks and in sleep.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):356-369. doi: 10.1016/j.bbagrm.2018.10.011. Epub 2018 Oct 31. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30391332 Review.
-
Editing of messenger RNA precursors and of tRNAs by adenosine to inosine conversion.FEBS Lett. 1999 Jun 4;452(1-2):71-6. doi: 10.1016/s0014-5793(99)00590-6. FEBS Lett. 1999. PMID: 10376681 Review.
Cited by
-
RNA editing and alternative splicing of the insect nAChR subunit alpha6 transcript: evolutionary conservation, divergence and regulation.BMC Evol Biol. 2007 Jun 27;7:98. doi: 10.1186/1471-2148-7-98. BMC Evol Biol. 2007. PMID: 17597521 Free PMC article.
-
Interplay between A-to-I Editing and Splicing of RNA: A Potential Point of Application for Cancer Therapy.Int J Mol Sci. 2022 May 8;23(9):5240. doi: 10.3390/ijms23095240. Int J Mol Sci. 2022. PMID: 35563631 Free PMC article. Review.
-
Dissecting the splicing mechanism of the Drosophila editing enzyme; dADAR.Nucleic Acids Res. 2009 Apr;37(5):1663-71. doi: 10.1093/nar/gkn1080. Epub 2009 Jan 19. Nucleic Acids Res. 2009. Retraction in: Nucleic Acids Res. 2024 May 22;52(9):5421. doi: 10.1093/nar/gkae221. PMID: 19153139 Free PMC article. Retracted.
-
Splicing and transcription touch base: co-transcriptional spliceosome assembly and function.Nat Rev Mol Cell Biol. 2017 Oct;18(10):637-650. doi: 10.1038/nrm.2017.63. Epub 2017 Aug 9. Nat Rev Mol Cell Biol. 2017. PMID: 28792005 Free PMC article. Review.
-
What do editors do? Understanding the physiological functions of A-to-I RNA editing by adenosine deaminase acting on RNAs.Open Biol. 2020 Jul;10(7):200085. doi: 10.1098/rsob.200085. Epub 2020 Jul 1. Open Biol. 2020. PMID: 32603639 Free PMC article. Review.
References
-
- Baskaran R, Escobar SR, Wang JY (1999) Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine (CTD)-tyrosine kinase-specific for the mammalian RNA polymerase II: possible role in transcription elongation. Cell Growth Differ 10: 387–396 - PubMed
-
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

