Identification of spliced gammaherpesvirus 68 LANA and v-cyclin transcripts and analysis of their expression in vivo during latent infection
- PMID: 16439562
- PMCID: PMC1367133
- DOI: 10.1128/JVI.80.4.2055-2062.2006
Identification of spliced gammaherpesvirus 68 LANA and v-cyclin transcripts and analysis of their expression in vivo during latent infection
Abstract
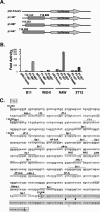
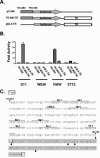
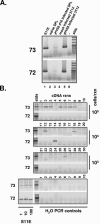
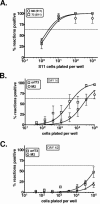
Regulation of orf73 (LANA) gene expression is critical to the establishment and maintenance of latency following infection by members of the gamma-2 herpesvirus (rhadinovirus) family. Previous studies of murine gammaherpesvirus 68 (gammaHV68) have demonstrated that loss of LANA function results in a complete failure to establish virus latency in the spleens of laboratory mice. Here we report the characterization of alternatively spliced LANA and v-cyclin (orf72) transcripts encoded by gammaHV68. Similar to other rhadinoviruses, alternative splicing, coupled with alternative 3' processing, of a ca. 16-kb transcriptional unit can lead to expression of either LANA or v-cyclin during gammaHV68 infection. Spliced LANA and v-cyclin transcripts were initially identified from an analysis of the gammaHV68 latently infected B-cell lymphoma cell line S11E, but were also detected during lytic infection of NIH 3T12 fibroblasts. 5' Random amplification of cDNA ends (RACE) analyses identified two distinct promoters, p1 and p2, that drive expression of spliced LANA transcripts. Analysis of p1 and p2, using transiently transfected reporter constructs, mapped the minimal sequences required for promoter activity and demonstrated that both promoters are active in the absence of any viral antigens. Analysis of spliced LANA and v-cyclin transcripts in spleens recovered from latently infected mice at days 16 and 42 postinfection revealed that spliced v-cyclin transcripts can only be detected sporadically, suggesting that these may be associated with cells reactivating from latency. In contrast, spliced LANA transcripts were detected in ca. 1 in 4,000 splenocytes harvested at day 16 postinfection. Notably, based on the frequency of viral genome-positive splenocytes at day 16 postinfection (ca. 1 in 200), only 5 to 10% of viral genome-positive splenocytes express LANA. The failure of the majority of infected splenocytes at day 16 postinfection to express LANA may contribute to the contraction in the frequency of latently infected splenocytes as chronic infection is established, due to failure to maintain the viral episome in proliferating B cells.
Figures

Similar articles
-
Identification of an Rta responsive promoter involved in driving gammaHV68 v-cyclin expression during virus replication.Virology. 2007 Sep 1;365(2):250-9. doi: 10.1016/j.virol.2007.03.021. Epub 2007 May 2. Virology. 2007. PMID: 17477952 Free PMC article.
-
The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency.J Virol. 2000 Aug;74(16):7451-61. doi: 10.1128/jvi.74.16.7451-7461.2000. J Virol. 2000. PMID: 10906198 Free PMC article.
-
The gammaherpesvirus 68 viral cyclin facilitates expression of LANA.PLoS Pathog. 2021 Nov 15;17(11):e1010019. doi: 10.1371/journal.ppat.1010019. eCollection 2021 Nov. PLoS Pathog. 2021. PMID: 34780571 Free PMC article.
-
Murine herpesvirus pathogenesis: a model for the analysis of molecular mechanisms of human gamma herpesvirus infections.Acta Microbiol Immunol Hung. 2005;52(1):41-71. doi: 10.1556/AMicr.52.2005.1.2. Acta Microbiol Immunol Hung. 2005. PMID: 15957234 Review.
-
Gamma interferon blocks gammaherpesvirus reactivation from latency in a cell type-specific manner.J Virol. 2007 Jun;81(11):6134-40. doi: 10.1128/JVI.00108-07. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360749 Free PMC article. Review.
Cited by
-
Relevance of BET Family Proteins in SARS-CoV-2 Infection.Biomolecules. 2021 Jul 30;11(8):1126. doi: 10.3390/biom11081126. Biomolecules. 2021. PMID: 34439792 Free PMC article. Review.
-
Identification of an Rta responsive promoter involved in driving gammaHV68 v-cyclin expression during virus replication.Virology. 2007 Sep 1;365(2):250-9. doi: 10.1016/j.virol.2007.03.021. Epub 2007 May 2. Virology. 2007. PMID: 17477952 Free PMC article.
-
Use of a virus-encoded enzymatic marker reveals that a stable fraction of memory B cells expresses latency-associated nuclear antigen throughout chronic gammaherpesvirus infection.J Virol. 2010 Aug;84(15):7523-34. doi: 10.1128/JVI.02572-09. Epub 2010 May 19. J Virol. 2010. PMID: 20484501 Free PMC article.
-
Viral latency and its regulation: lessons from the gamma-herpesviruses.Cell Host Microbe. 2010 Jul 22;8(1):100-15. doi: 10.1016/j.chom.2010.06.014. Cell Host Microbe. 2010. PMID: 20638646 Free PMC article. Review.
-
Tiled microarray identification of novel viral transcript structures and distinct transcriptional profiles during two modes of productive murine gammaherpesvirus 68 infection.J Virol. 2012 Apr;86(8):4340-57. doi: 10.1128/JVI.05892-11. Epub 2012 Feb 8. J Virol. 2012. PMID: 22318145 Free PMC article.
References
-
- Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. - PubMed
-
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

