Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities
- PMID: 16439523
- PMCID: PMC1367131
- DOI: 10.1128/JVI.80.4.1662-1671.2006
Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities
Abstract
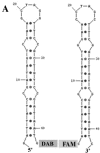
Poliovirus protein 3AB displayed nucleic acid chaperone activity in promoting the hybridization of complementary nucleic acids and destabilizing secondary structure. Hybridization reactions at 30 degrees C between 20- and 40-nucleotide RNA oligonucleotides and 179- or 765-nucleotide RNAs that contained a complementary region were greatly enhanced in the presence of 3AB. The effect was nonspecific as reactions between DNA oligonucleotides and RNA or DNA templates were also enhanced. Reactions were optimal with 1 mM MgCl(2) and 20 mM KCl. Analysis of the reactions with various 3AB and template concentrations indicated that enhancement required a critical amount of 3AB that increased as the concentration of nucleic acid increased. This was consistent with a requirement for 3AB to "coat" the nucleic acids for enhancement. The helix-destabilizing activity of 3AB was tested in an assay with two 42-nucleotide completely complementary DNAs. Each complement formed a strong stem-loop (DeltaG = -7.2 kcal/mol) that required unwinding for hybridization to occur. DNAs were modified at the 3' or 5' end with fluorescent probes such that hybridization resulted in quenching of the fluorescent signal. Under optimal conditions at 30 degrees C, 3AB stimulated hybridization in a concentration-dependent manner, as did human immunodeficiency virus nucleocapsid protein, an established chaperone. The results are discussed with respect to the role of 3AB in viral replication and recombination.
Figures





Similar articles
-
The twenty-nine amino acid C-terminal cytoplasmic domain of poliovirus 3AB is critical for nucleic acid chaperone activity.RNA Biol. 2010 Nov-Dec;7(6):820-9. doi: 10.4161/rna.7.6.13781. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045553 Free PMC article.
-
HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity.J Mol Biol. 2002 May 3;318(3):749-64. doi: 10.1016/S0022-2836(02)00043-8. J Mol Biol. 2002. PMID: 12054820
-
The identification and characterization of nucleic acid chaperone activity of human enterovirus 71 nonstructural protein 3AB.Virology. 2014 Sep;464-465:353-364. doi: 10.1016/j.virol.2014.07.037. Epub 2014 Aug 9. Virology. 2014. PMID: 25113906 Free PMC article.
-
DNA probes: applications of the principles of nucleic acid hybridization.Crit Rev Biochem Mol Biol. 1991;26(3-4):227-59. doi: 10.3109/10409239109114069. Crit Rev Biochem Mol Biol. 1991. PMID: 1718662 Review.
-
Nucleic acid encoding to program self-assembly in chemical biology.Chem Soc Rev. 2008 Jul;37(7):1330-6. doi: 10.1039/b706610b. Epub 2008 Apr 17. Chem Soc Rev. 2008. PMID: 18568159 Review.
Cited by
-
Picornaviruses: A View from 3A.Viruses. 2021 Mar 11;13(3):456. doi: 10.3390/v13030456. Viruses. 2021. PMID: 33799649 Free PMC article. Review.
-
Correlation between recombination junctions and RNA secondary structure elements in poliovirus Sabin strains.Virus Genes. 2010 Oct;41(2):181-91. doi: 10.1007/s11262-010-0512-5. Epub 2010 Jul 17. Virus Genes. 2010. PMID: 20640496 Free PMC article.
-
The nonstructural protein 2C of a Picorna-like virus displays nucleic acid helix destabilizing activity that can be functionally separated from its ATPase activity.J Virol. 2013 May;87(9):5205-18. doi: 10.1128/JVI.00245-13. Epub 2013 Feb 28. J Virol. 2013. PMID: 23449794 Free PMC article.
-
Initiation of protein-primed picornavirus RNA synthesis.Virus Res. 2015 Aug 3;206:12-26. doi: 10.1016/j.virusres.2014.12.028. Epub 2015 Jan 12. Virus Res. 2015. PMID: 25592245 Free PMC article. Review.
-
Expanding knowledge of P3 proteins in the poliovirus lifecycle.Future Microbiol. 2010 Jun;5(6):867-81. doi: 10.2217/fmb.10.40. Future Microbiol. 2010. PMID: 20521933 Free PMC article. Review.
References
-
- Arnold, J. J., S. K. Ghosh, and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol). J. Biol. Chem. 274:37060-37069. - PubMed
-
- Banerjee, R., and A. Dasgupta. 2001. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 82:2621-2627. - PubMed
-
- Baron, M. H., and D. Baltimore. 1982. Anti-VPg antibody inhibition of the poliovirus replicase reaction and production of covalent complexes of VPg-related proteins and RNA. Cell 30:745-752. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

