A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network
- PMID: 16432214
- PMCID: PMC1360546
- DOI: 10.1073/pnas.0508103103
A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network
Abstract
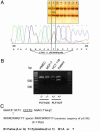
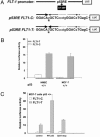
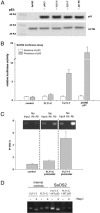
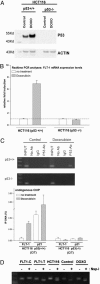
The VEGF system is essential for angiogenesis. VEGF overexpression frequently correlates with increased microvascularity and metastasis and decreased spontaneous apoptosis. Although a precise mechanism has not been established, studies suggest that VEGF expression is negatively regulated by p53, a master regulator and tumor suppressor. There are no reports of additional components of the VEGF signal transduction pathway being part of the p53 transcriptional network. A target of VEGF, the VEGF receptor 1/flt-1, can regulate growth and migration of endothelial cells and modulate angiogenesis. VEGF appears to be up-regulated in various cancers in which flt-1 may have a role in tumor progression and metastasis. We identified a C-to-T SNP upstream of the transcriptional start site in approximately 6% of the people examined. The SNP is located within a putative p53 response element. Only the promoter with the T SNP (FLT1-T) was responsive to p53 when examined with reporter assays or by endogenous gene expression analysis in cell lines with different SNP status. In response to doxorubicin-induced DNA damage, there was clear allele discrimination based on p53 binding at the FLT1-T but not FLT1-C promoters as well as p53-dependent induction of flt-1 mRNA, which required the presence of FLT1-T. Our results establish that p53 can differentially stimulate transcription at a polymorphic variant of the flt-1 promoter and directly places the VEGF system in the p53 stress-response network via flt-1 in a significant fraction of the human population. We suggest that the p53-VEGF-flt-1 interaction is relevant to risks in angiogenesis-associated diseases, including cancer.
Figures
Similar articles
-
p53-dependent inhibition of progestin-induced VEGF expression in human breast cancer cells.J Steroid Biochem Mol Biol. 2005 Feb;93(2-5):173-82. doi: 10.1016/j.jsbmb.2004.12.011. Epub 2005 Jan 28. J Steroid Biochem Mol Biol. 2005. PMID: 15860260
-
Decoy receptor 2 (DcR2) is a p53 target gene and regulates chemosensitivity.Cancer Res. 2005 Oct 15;65(20):9169-75. doi: 10.1158/0008-5472.CAN-05-0939. Cancer Res. 2005. PMID: 16230375
-
Histone deacetylase 5 is not a p53 target gene, but its overexpression inhibits tumor cell growth and induces apoptosis.Cancer Res. 2002 May 15;62(10):2913-22. Cancer Res. 2002. PMID: 12019172
-
Transcriptional activities of mutant p53: when mutations are more than a loss.J Cell Biochem. 2004 Nov 15;93(5):878-86. doi: 10.1002/jcb.20271. J Cell Biochem. 2004. PMID: 15449312 Review.
-
Regulation of the p53 transcriptional activity.J Cell Biochem. 2006 Feb 15;97(3):448-58. doi: 10.1002/jcb.20700. J Cell Biochem. 2006. PMID: 16288459 Review.
Cited by
-
Phenotype specific analyses reveal distinct regulatory mechanism for chronically activated p53.PLoS Genet. 2015 Mar 19;11(3):e1005053. doi: 10.1371/journal.pgen.1005053. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25790137 Free PMC article.
-
Transcriptional functionality of germ line p53 mutants influences cancer phenotype.Clin Cancer Res. 2007 Jul 1;13(13):3789-95. doi: 10.1158/1078-0432.CCR-06-2545. Clin Cancer Res. 2007. PMID: 17606709 Free PMC article.
-
Estrogen receptor acting in cis enhances WT and mutant p53 transactivation at canonical and noncanonical p53 target sequences.Proc Natl Acad Sci U S A. 2010 Jan 26;107(4):1500-5. doi: 10.1073/pnas.0909129107. Epub 2010 Jan 4. Proc Natl Acad Sci U S A. 2010. PMID: 20080630 Free PMC article.
-
Algorithm for prediction of tumour suppressor p53 affinity for binding sites in DNA.Nucleic Acids Res. 2008 Mar;36(5):1589-98. doi: 10.1093/nar/gkm1040. Epub 2008 Jan 30. Nucleic Acids Res. 2008. PMID: 18234719 Free PMC article.
-
Functional evolution of the p53 regulatory network through its target response elements.Proc Natl Acad Sci U S A. 2008 Jan 22;105(3):944-9. doi: 10.1073/pnas.0704694105. Epub 2008 Jan 10. Proc Natl Acad Sci U S A. 2008. PMID: 18187580 Free PMC article.
References
-
- Luttun, A., Autiero, M., Tjwa, M. & Carmeliet, P. (2004) Biochim. Biophys. Acta 1654, 79-94. - PubMed
-
- Hoeben, A., Landuyt, B., Highley, M. S., Wildiers, H., Van Oosterom, A. T. & De Bruijn, E. A. (2004) Pharmacol. Rev. 56, 549-580. - PubMed
-
- Bellamy, W. T. (2002) Curr. Opin. Oncol. 14, 649-656. - PubMed
-
- Ferrara, N. (2004) Endocr. Rev. 25, 581-611. - PubMed
-
- Shibuya, M. (2001) Cell. Struct. Funct. 26, 25-35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

