Eleven ancestral gene families lost in mammals and vertebrates while otherwise universally conserved in animals
- PMID: 16420703
- PMCID: PMC1382263
- DOI: 10.1186/1471-2148-6-5
Eleven ancestral gene families lost in mammals and vertebrates while otherwise universally conserved in animals
Abstract
Background: Gene losses played a role which may have been as important as gene and genome duplications and rearrangements, in modelling today species' genomes from a common ancestral set of genes. The set and diversity of protein-coding genes in a species has direct output at the functional level. While gene losses have been reported in all the major lineages of the metazoan tree of life, none have proposed a focus on specific losses in the vertebrates and mammals lineages. In contrast, genes lost in protostomes (i.e. arthropods and nematodes) but still present in vertebrates have been reported and extensively detailed. This probable over-anthropocentric way of comparing genomes does not consider as an important phenomena, gene losses in species that are usually described as "higher". However reporting universally conserved genes throughout evolution that have recently been lost in vertebrates and mammals could reveal interesting features about the evolution of our genome, particularly if these losses can be related to losses of capability.
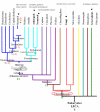
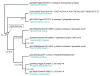
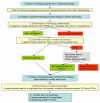
Results: We report 11 gene families conserved throughout eukaryotes from yeasts (such as Saccharomyces cerevisiae) to bilaterian animals (such as Drosophila melanogaster or Caenorhabditis elegans). This evolutionarily wide conservation suggests they were present in the last common ancestors of fungi and metazoan animals. None of these 11 gene families are found in human nor mouse genomes, and their absence generally extends to all vertebrates. A total of 8 out of these 11 gene families have orthologs in plants, suggesting they were present in the Last Eukaryotic Common Ancestor (LECA). We investigated known functional information for these 11 gene families. This allowed us to correlate some of the lost gene families to loss of capabilities.
Conclusion: Mammalian and vertebrate genomes lost evolutionary conserved ancestral genes that are probably otherwise not dispensable in eukaryotes. Hence, the human genome, which is generally viewed as being the result of increased complexity and gene-content, has also evolved through simplification and gene losses. This acknowledgement confirms, as already suggested, that the genome of our far ancestor was probably more complex than ever considered.
Figures
Similar articles
-
A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes.Genome Biol. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. Epub 2004 Jan 15. Genome Biol. 2004. PMID: 14759257 Free PMC article.
-
Correlating traits of gene retention, sequence divergence, duplicability and essentiality in vertebrates, arthropods, and fungi.Genome Biol Evol. 2011;3:75-86. doi: 10.1093/gbe/evq083. Epub 2010 Dec 9. Genome Biol Evol. 2011. PMID: 21148284 Free PMC article.
-
The Natural History of Teneurins: A Billion Years of Evolution in Three Key Steps.Front Neurosci. 2019 Mar 15;13:109. doi: 10.3389/fnins.2019.00109. eCollection 2019. Front Neurosci. 2019. PMID: 30930727 Free PMC article.
-
Origin and evolution of spliceosomal introns.Biol Direct. 2012 Apr 16;7:11. doi: 10.1186/1745-6150-7-11. Biol Direct. 2012. PMID: 22507701 Free PMC article. Review.
-
GnRH receptors and peptides: skating backward.Gen Comp Endocrinol. 2014 Dec 1;209:118-34. doi: 10.1016/j.ygcen.2014.07.025. Epub 2014 Aug 5. Gen Comp Endocrinol. 2014. PMID: 25107740 Review.
Cited by
-
Evolution by gene loss.Nat Rev Genet. 2016 Jul;17(7):379-91. doi: 10.1038/nrg.2016.39. Epub 2016 Apr 18. Nat Rev Genet. 2016. PMID: 27087500 Review.
-
Fasting induces CART down-regulation in the zebrafish nervous system in a cannabinoid receptor 1-dependent manner.Mol Endocrinol. 2012 Aug;26(8):1316-26. doi: 10.1210/me.2011-1180. Epub 2012 Jun 14. Mol Endocrinol. 2012. PMID: 22700585 Free PMC article.
-
Functional Insights From the Evolutionary Diversification of Big Defensins.Front Immunol. 2020 Apr 30;11:758. doi: 10.3389/fimmu.2020.00758. eCollection 2020. Front Immunol. 2020. PMID: 32425943 Free PMC article. Review.
-
Metazoan Hsp70-based protein disaggregases: emergence and mechanisms.Front Mol Biosci. 2015 Oct 9;2:57. doi: 10.3389/fmolb.2015.00057. eCollection 2015. Front Mol Biosci. 2015. PMID: 26501065 Free PMC article. Review.
-
Mechanisms of Gene Duplication and Translocation and Progress towards Understanding Their Relative Contributions to Animal Genome Evolution.Int J Evol Biol. 2012;2012:846421. doi: 10.1155/2012/846421. Epub 2012 Aug 7. Int J Evol Biol. 2012. PMID: 22919542 Free PMC article.
References
-
- Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Rogozin IB, Smirnov S, Sorokin AV, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

