Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development
- PMID: 16418395
- PMCID: PMC2118073
- DOI: 10.1084/jem.20051645
Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development
Abstract
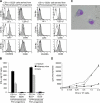
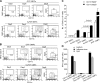
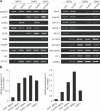
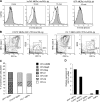
Flt3 ligand (Flt3L) is a nonredundant cytokine in type I interferon-producing cell (IPC) and dendritic cell (DC) development, and IPC and DC differentiation potential is confined to Flt3+ hematopoietic progenitor cells. Here, we show that overexpression of human Flt3 in Flt3- (Flt3(-)Lin(-)IL-7Ralpha(-)Thy1.1(-)c-Kit+) and Flt3+ (Flt3(+)Lin(-)IL-7Ralpha(-)Thy1.1(-)c-Kit+) hematopoietic progenitors rescues and enhances their IPC and DC differentiation potential, respectively. In defined hematopoietic cell populations, such as Flt3- megakaryocyte/erythrocyte-restricted progenitors (MEPs), enforced Flt3 signaling induces transcription of IPC, DC, and granulocyte/macrophage (GM) development-affiliated genes, including STAT3, PU.1, and G-/M-/GM-CSFR, and activates differentiation capacities to these lineages. Moreover, ectopic expression of Flt3 downstream transcription factors STAT3 or PU.1 in Flt3- MEPs evokes Flt3 receptor expression and instructs differentiation into IPCs, DCs, and myelomonocytic cells, whereas GATA-1 expression and consecutive megakaryocyte/erythrocyte development is suppressed. Based on these data, we propose a demand-regulated, cytokine-driven DC and IPC regeneration model, in which high Flt3L levels initiate a self-sustaining, Flt3-STAT3- and Flt3-PU.1-mediated IPC and DC differentiation program in Flt3+ hematopoietic progenitor cells.
Figures


Similar articles
-
Flt3 in regulation of type I interferon-producing cell and dendritic cell development.Ann N Y Acad Sci. 2007 Jun;1106:253-61. doi: 10.1196/annals.1392.015. Epub 2007 Mar 14. Ann N Y Acad Sci. 2007. PMID: 17360795 Review.
-
Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow.Nat Immunol. 2007 Nov;8(11):1207-16. doi: 10.1038/ni1518. Epub 2007 Oct 7. Nat Immunol. 2007. PMID: 17922016
-
Differentiation of NK1.1+, Ly49+ NK cells from flt3+ multipotent marrow progenitor cells.J Immunol. 1999 Sep 1;163(5):2648-56. J Immunol. 1999. PMID: 10453005
-
Dendritic cell lineage commitment is instructed by distinct cytokine signals.Eur J Cell Biol. 2012 Jun-Jul;91(6-7):515-23. doi: 10.1016/j.ejcb.2011.09.007. Epub 2011 Nov 9. Eur J Cell Biol. 2012. PMID: 22078373
-
Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells.Leukemia. 1996 Apr;10(4):588-99. Leukemia. 1996. PMID: 8618433 Review.
Cited by
-
HIV gag protein is efficiently cross-presented when targeted with an antibody towards the DEC-205 receptor in Flt3 ligand-mobilized murine DC.Eur J Immunol. 2010 Jan;40(1):36-46. doi: 10.1002/eji.200939748. Eur J Immunol. 2010. PMID: 19830741 Free PMC article.
-
Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling.Genome Biol. 2008 Jan 24;9(1):R17. doi: 10.1186/gb-2008-9-1-r17. Genome Biol. 2008. PMID: 18218067 Free PMC article.
-
Signal transduction inhibition of APCs diminishes th17 and Th1 responses in experimental autoimmune encephalomyelitis.J Immunol. 2009 Apr 1;182(7):4192-9. doi: 10.4049/jimmunol.0803631. J Immunol. 2009. PMID: 19299717 Free PMC article.
-
The Cytokine Flt3-Ligand in Normal and Malignant Hematopoiesis.Int J Mol Sci. 2017 May 24;18(6):1115. doi: 10.3390/ijms18061115. Int J Mol Sci. 2017. PMID: 28538663 Free PMC article. Review.
-
Sensitivity of Dendritic Cells to Microenvironment Signals.J Immunol Res. 2016;2016:4753607. doi: 10.1155/2016/4753607. Epub 2016 Mar 21. J Immunol Res. 2016. PMID: 27088097 Free PMC article. Review.
References
-
- Kondo, M., A.J. Wagers, M.G. Manz, S.S. Prohaska, D.C. Scherer, G.F. Beilhack, J.A. Shizuru, and I.L. Weissman. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806. - PubMed
-
- Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. - PubMed
-
- Galy, A., M. Travis, D. Cen, and B. Chen. 1995. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 3:459–473. - PubMed
-
- Kondo, M., I.L. Weissman, and K. Akashi. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

