Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway
- PMID: 16402859
- PMCID: PMC1334386
- DOI: 10.1371/journal.pbio.0040027
Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway
Abstract
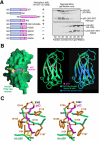
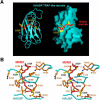
Herpesvirus-associated ubiquitin-specific protease (HAUSP, also known as USP7), a deubiquitylating enzyme of the ubiquitin-specific processing protease family, specifically deubiquitylates both p53 and MDM2, hence playing an important yet enigmatic role in the p53-MDM2 pathway. Here we demonstrate that both p53 and MDM2 specifically recognize the N-terminal tumor necrosis factor-receptor associated factor (TRAF)-like domain of HAUSP in a mutually exclusive manner. HAUSP preferentially forms a stable HAUSP-MDM2 complex even in the presence of excess p53. The HAUSP-binding elements were mapped to a peptide fragment in the carboxy-terminus of p53 and to a short-peptide region preceding the acidic domain of MDM2. The crystal structures of the HAUSP TRAF-like domain in complex with p53 and MDM2 peptides, determined at 2.3-A and 1.7-A resolutions, respectively, reveal that the MDM2 peptide recognizes the same surface groove in HAUSP as that recognized by p53 but mediates more extensive interactions. Structural comparison led to the identification of a consensus peptide-recognition sequence by HAUSP. These results, together with the structure of a combined substrate-binding-and-deubiquitylation domain of HAUSP, provide important insights into regulation of the p53-MDM2 pathway by HAUSP.
Figures


Comment in
-
Structural insights into the regulation of a key tumor suppressor.PLoS Biol. 2006 Feb;4(2):e40. doi: 10.1371/journal.pbio.0040040. Epub 2006 Jan 17. PLoS Biol. 2006. PMID: 20076526 Free PMC article. No abstract available.
Similar articles
-
Molecular recognition of p53 and MDM2 by USP7/HAUSP.Nat Struct Mol Biol. 2006 Mar;13(3):285-91. doi: 10.1038/nsmb1067. Epub 2006 Feb 12. Nat Struct Mol Biol. 2006. PMID: 16474402
-
C-terminal region of USP7/HAUSP is critical for deubiquitination activity and contains a second mdm2/p53 binding site.Arch Biochem Biophys. 2010 Nov 15;503(2):207-12. doi: 10.1016/j.abb.2010.08.020. Epub 2010 Sep 15. Arch Biochem Biophys. 2010. PMID: 20816748
-
Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.Mol Cell. 2005 Apr 1;18(1):25-36. doi: 10.1016/j.molcel.2005.02.029. Mol Cell. 2005. PMID: 15808506
-
Modulation of the p53/MDM2 interplay by HAUSP inhibitors.J Mol Cell Biol. 2017 Feb 1;9(1):45-52. doi: 10.1093/jmcb/mjw049. J Mol Cell Biol. 2017. PMID: 27927749 Free PMC article. Review.
-
ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation.Cell Cycle. 2005 Sep;4(9):1166-70. doi: 10.4161/cc.4.9.1981. Epub 2005 Sep 29. Cell Cycle. 2005. PMID: 16082221 Review.
Cited by
-
Structural Characterization of Interaction between Human Ubiquitin-specific Protease 7 and Immediate-Early Protein ICP0 of Herpes Simplex Virus-1.J Biol Chem. 2015 Sep 18;290(38):22907-18. doi: 10.1074/jbc.M115.664805. Epub 2015 Jul 29. J Biol Chem. 2015. PMID: 26224631 Free PMC article.
-
Structural basis of ubiquitin recognition by the deubiquitinating protease USP2.Structure. 2006 Aug;14(8):1293-302. doi: 10.1016/j.str.2006.06.012. Structure. 2006. PMID: 16905103 Free PMC article.
-
Deubiquitinases: Pro-oncogenic Activity and Therapeutic Targeting in Blood Malignancies.Trends Immunol. 2020 Apr;41(4):327-340. doi: 10.1016/j.it.2020.02.004. Epub 2020 Mar 2. Trends Immunol. 2020. PMID: 32139316 Free PMC article. Review.
-
The role of ubiquitination in spinal and bulbar muscular atrophy.Front Mol Neurosci. 2022 Oct 6;15:1020143. doi: 10.3389/fnmol.2022.1020143. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36277484 Free PMC article. Review.
-
Deubiquitination in virus infection.Virology. 2007 Jun 5;362(2):245-56. doi: 10.1016/j.virol.2006.12.035. Epub 2007 Feb 7. Virology. 2007. PMID: 17291557 Free PMC article. Review.
References
-
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. - PubMed
-
- Michael D, Oren M. The p53-MDM2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. - PubMed
-
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. - PubMed
-
- Brooks CL, Gu W. Ubiquitylation, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. - PubMed
-
- Haupt Y, Robles AI, Prives C, Rotter V. Deconstruction of p53 functions and regulation. Oncogene. 2002;21:8223–8231. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

