Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine
- PMID: 16388603
- PMCID: PMC2519119
- DOI: 10.1021/bi052014t
Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine
Abstract
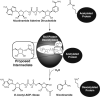
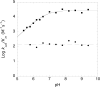
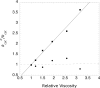
Sir2 NAD+-dependent protein deacetylases are implicated in a variety of cellular processes such as apoptosis, gene silencing, life-span regulation, and fatty acid metabolism. Despite this, there have been relatively few investigations into the detailed chemical mechanism. Sir2 proteins (sirtuins) catalyze the chemical conversion of NAD+ and acetylated lysine to nicotinamide, deacetylated lysine, and 2'-O-acetyl-ADP-ribose (OAADPr). In this study, Sir2-catalyzed reactions are shown to transfer an 18O label from the peptide acetyl group to the ribose 1'-position of OAADPr, providing direct evidence for the formation of a covalent alpha-1'-O-alkylamidate, whose existence is further supported by the observed methanolysis of the alpha-1'-O-alkylamidate intermediate to yield beta-1'-O-methyl-ADP-ribose in a Sir2 histidine-to-alanine mutant. This conserved histidine (His-135 in HST2) activates the ribose 2'-hydroxyl for attack on the alpha-1'-O-alkylamidate. The histidine mutant is stalled at the intermediate, allowing water and other alcohols to compete kinetically with the attacking 2'-hydroxyl. Measurement of the pH dependence of kcat and kcat/Km values for both wild-type and histidine-to-alanine mutant enzymes confirms roles of this residue in NAD+ binding and in general-base activation of the 2'-hydroxyl. Also, transfer of an 18O label from water to the carbonyl oxygen of the acetyl group in OAADPr is consistent with water addition to the proposed 1',2'-cyclic intermediate formed after 2'-hydroxyl attack on the alpha-1'-O-alkylamidate. The effect of pH and of solvent viscosity on the kcat values suggests that final product release is rate-limiting in the wild-type enzyme. Implications of this new evidence on the mechanisms of deacetylation and possible ADP-ribosylation catalyzed by Sir2 deacetylases are discussed.
Figures
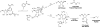





Similar articles
-
Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.Proc Natl Acad Sci U S A. 2004 Jun 8;101(23):8563-8. doi: 10.1073/pnas.0401057101. Epub 2004 May 18. Proc Natl Acad Sci U S A. 2004. PMID: 15150415 Free PMC article.
-
Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases.Biochemistry. 2004 Aug 3;43(30):9877-87. doi: 10.1021/bi049592e. Biochemistry. 2004. PMID: 15274642
-
Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry.Biochemistry. 2003 Aug 12;42(31):9249-56. doi: 10.1021/bi034959l. Biochemistry. 2003. PMID: 12899610
-
SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review.
-
The Sir 2 family of protein deacetylases.Curr Opin Chem Biol. 2005 Oct;9(5):431-40. doi: 10.1016/j.cbpa.2005.08.010. Curr Opin Chem Biol. 2005. PMID: 16122969 Review.
Cited by
-
Entamoeba histolytica sirtuin EhSir2a deacetylates tubulin and regulates the number of microtubular assemblies during the cell cycle.Cell Microbiol. 2010 Jul;12(7):1002-14. doi: 10.1111/j.1462-5822.2010.01449.x. Epub 2010 Feb 9. Cell Microbiol. 2010. PMID: 20148900 Free PMC article.
-
Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going?Microbiol Mol Biol Rev. 2006 Sep;70(3):789-829. doi: 10.1128/MMBR.00040-05. Microbiol Mol Biol Rev. 2006. PMID: 16959969 Free PMC article. Review.
-
Mechanisms and molecular probes of sirtuins.Chem Biol. 2008 Oct 20;15(10):1002-13. doi: 10.1016/j.chembiol.2008.09.009. Chem Biol. 2008. PMID: 18940661 Free PMC article. Review.
-
Sirtuin 1 and sirtuin 3: physiological modulators of metabolism.Physiol Rev. 2012 Jul;92(3):1479-514. doi: 10.1152/physrev.00022.2011. Physiol Rev. 2012. PMID: 22811431 Free PMC article. Review.
-
DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1.J Biol Chem. 2010 Apr 23;285(17):13223-32. doi: 10.1074/jbc.M110.102574. Epub 2010 Feb 18. J Biol Chem. 2010. PMID: 20167603 Free PMC article.
References
-
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. - PubMed
-
- Gasser SM, Cockell MM. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. - PubMed
-
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. - PubMed
-
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

