Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding
- PMID: 16382153
- PMCID: PMC1346915
- DOI: 10.1128/MCB.26.2.630-642.2006
Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding
Abstract
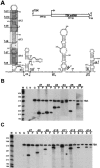
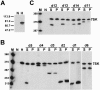
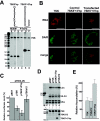
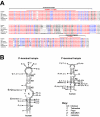
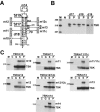
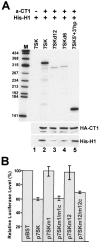
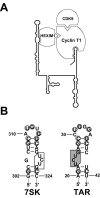
The positive transcription elongation factor b (P-TEFb), a complex of Cdk9 and cyclin T1/T2, stimulates transcription by phosphorylating RNA polymerase II. The 7SK small nuclear RNA, in cooperation with HEXIM1 protein, functions as a general polymerase II transcription regulator by sequestering P-TEFb into a large kinase-inactive 7SK/HEXIM1/P-TEFb complex. Here, determination and characterization of the functionally essential elements of human 7SK snRNA directing HEXIM1 and P-TEFb binding led to a new model for the assembly of the 7SK/HEXIM1/P-TEFb regulatory complex. We demonstrate that two structurally and functionally distinct protein binding elements located in the 5'- and 3'-terminal hairpins of 7SK support the in vivo recruitment of HEXIM1 and P-TEFb. Consistently, a minimal regulatory RNA composed of the 5' and 3' hairpins of 7SK can modulate polymerase II transcription in HeLa cells. HEXIM1 binds independently and specifically to the G24-C48/G60-C87 distal segment of the 5' hairpin of 7SK. Binding of HEXIM1 is a prerequisite for association of P-TEFb with the G302-C324 apical region of the 3' hairpin of 7SK that is highly reminiscent of the human immunodeficiency virus transactivation-responsive RNA.
Figures

Similar articles
-
A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb.Mol Cell Biol. 2004 Jun;24(12):5094-105. doi: 10.1128/MCB.24.12.5094-5105.2004. Mol Cell Biol. 2004. PMID: 15169877 Free PMC article.
-
Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA.Mol Cell. 2003 Oct;12(4):971-82. doi: 10.1016/s1097-2765(03)00388-5. Mol Cell. 2003. PMID: 14580347
-
Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb.EMBO J. 2007 Aug 8;26(15):3570-80. doi: 10.1038/sj.emboj.7601783. Epub 2007 Jul 5. EMBO J. 2007. PMID: 17611602 Free PMC article.
-
7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor.RNA Biol. 2009 Apr-Jun;6(2):122-8. doi: 10.4161/rna.6.2.8115. Epub 2009 Apr 6. RNA Biol. 2009. PMID: 19246988 Review.
-
P-TEFb: The master regulator of transcription elongation.Mol Cell. 2023 Feb 2;83(3):393-403. doi: 10.1016/j.molcel.2022.12.006. Epub 2023 Jan 3. Mol Cell. 2023. PMID: 36599353 Free PMC article. Review.
Cited by
-
hLARP7 C-terminal domain contains an xRRM that binds the 3' hairpin of 7SK RNA.Nucleic Acids Res. 2016 Nov 16;44(20):9977-9989. doi: 10.1093/nar/gkw833. Epub 2016 Sep 26. Nucleic Acids Res. 2016. PMID: 27679474 Free PMC article.
-
CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation.Cell Cycle. 2010 May;9(9):1697-705. doi: 10.4161/cc.9.9.11346. Epub 2010 May 29. Cell Cycle. 2010. PMID: 20436276 Free PMC article. Review.
-
The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes.Mol Cell Biol. 2007 Oct;27(20):6996-7006. doi: 10.1128/MCB.00975-07. Epub 2007 Aug 20. Mol Cell Biol. 2007. PMID: 17709395 Free PMC article.
-
Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA.Mol Cell Biol. 2009 Mar;29(5):1375-87. doi: 10.1128/MCB.01365-08. Epub 2008 Dec 22. Mol Cell Biol. 2009. PMID: 19103749 Free PMC article.
-
Reconstitution of a functional 7SK snRNP.Nucleic Acids Res. 2017 Jun 20;45(11):6864-6880. doi: 10.1093/nar/gkx262. Nucleic Acids Res. 2017. PMID: 28431135 Free PMC article.
References
-
- Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. - PubMed
-
- Brasier, A. R., and J. J. Fortin. 1994. Nonisotopic assays for reporter gene activity, p. 9.7.12-9.7.21. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
-
- Byers, S. A., J. P. Price, J. J. Cooper, Q. Li, and D. H. Price. 2005. HEXIM2, a HEXIM1 related protein, regulates P-TEFb through association with 7SK. J. Biol. Chem. 280:16360-16367. - PubMed
-
- Chao, S.-H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
