GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences
- PMID: 16381828
- PMCID: PMC1347436
- DOI: 10.1093/nar/gkj073
GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences
Abstract
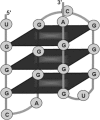
Guanine-rich nucleic acids are known to form highly stable G-quadruplex structures, also known as G-quartets. Recently, there has been a tremendous amount of interest in studying G-quadruplexes owing to the realization of their biological importance. G-rich sequences (GRSs) capable of forming G-quadruplexes are found in the vicinity of polyadenylation regions and are involved in regulating 3' end processing of mammalian pre-mRNAs. G-rich motifs are also known to play an important role in alternative, tissue-specific splicing by interacting with hnRNP H protein subfamily. Whether quadruplex structure directly plays a role in regulating RNA processing events requires further investigation. To date there has not been a comprehensive effort to study G-quadruplexes near RNA processing sites. We have applied a computational approach to map putative Quadruplex forming GRSs within the transcribed regions of a large number of alternatively processed human and mouse gene sequences that were obtained as fully annotated entries from GenBank and RefSeq. We have used the computed data to build the GRSDB database that provides a unique avenue for studying G-quadruplexes in the context of RNA processing sites. GRSDB website offers visual comparison of G-quadruplex distribution patterns among all the alternative RNA products of a gene with the help of dynamic graphics. At present, GRSDB contains data from 1310 human and mouse genes, of which 1188 are alternatively processed. It has a total of 379,223 predicted G-quadruplexes, of which 54,252 are near RNA processing sites. GRSDB is a good resource for researchers interested in investigating the functional relevance of G-quadruplexes, especially in the context of alternative RNA processing. It can be accessed at http://bioinformatics.ramapo.edu/grsdb/.
Figures


Similar articles
-
QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W676-82. doi: 10.1093/nar/gkl253. Nucleic Acids Res. 2006. PMID: 16845096 Free PMC article.
-
GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs.Nucleic Acids Res. 2008 Jan;36(Database issue):D141-8. doi: 10.1093/nar/gkm982. Epub 2007 Nov 27. Nucleic Acids Res. 2008. PMID: 18045785 Free PMC article.
-
G-quadruplex formation of FXYD1 pre-mRNA indicates the possibility of regulating expression of its protein product.Arch Biochem Biophys. 2014 Oct 15;560:52-8. doi: 10.1016/j.abb.2014.07.016. Epub 2014 Jul 19. Arch Biochem Biophys. 2014. PMID: 25051342
-
Biological aspects of DNA/RNA quadruplexes.Biopolymers. 2000-2001;56(3):209-27. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. Biopolymers. 2000. PMID: 11745112 Review.
-
G-quadruplexes in RNA biology.Wiley Interdiscip Rev RNA. 2012 Jul-Aug;3(4):495-507. doi: 10.1002/wrna.1113. Epub 2012 Apr 4. Wiley Interdiscip Rev RNA. 2012. PMID: 22488917 Review.
Cited by
-
Discovery of widespread GTP-binding motifs in genomic DNA and RNA.Chem Biol. 2013 Apr 18;20(4):521-32. doi: 10.1016/j.chembiol.2013.02.015. Chem Biol. 2013. PMID: 23601641 Free PMC article.
-
Experimental approaches to identify cellular G-quadruplex structures and functions.Methods. 2012 May;57(1):84-92. doi: 10.1016/j.ymeth.2012.01.008. Epub 2012 Feb 11. Methods. 2012. PMID: 22343041 Free PMC article.
-
RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms.J Mol Biol. 2017 Jul 7;429(14):2127-2147. doi: 10.1016/j.jmb.2017.05.017. Epub 2017 May 26. J Mol Biol. 2017. PMID: 28554731 Free PMC article. Review.
-
Quadfinder: server for identification and analysis of quadruplex-forming motifs in nucleotide sequences.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W683-5. doi: 10.1093/nar/gkl299. Nucleic Acids Res. 2006. PMID: 16845097 Free PMC article.
-
G-quadruplexes: the beginning and end of UTRs.Nucleic Acids Res. 2008 Nov;36(19):6260-8. doi: 10.1093/nar/gkn511. Epub 2008 Oct 2. Nucleic Acids Res. 2008. PMID: 18832370 Free PMC article.

