Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication
- PMID: 16352554
- PMCID: PMC1317528
- DOI: 10.1128/JVI.80.1.296-305.2006
Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication
Abstract
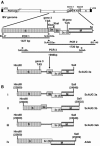
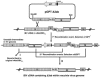
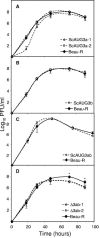
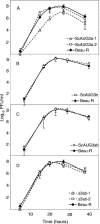
Gene 3 of infectious bronchitis virus is tricistronic; open reading frames (ORFs) 3a and 3b encode two small nonstructural (ns) proteins, 3a and 3b, of unknown function, and a third, structural protein E, is encoded by ORF 3c. To determine if either the 3a or the 3b protein is required for replication, we first modified their translation initiation codons to prevent translation of the 3a and 3b proteins from recombinant infectious bronchitis viruses (rIBVs). Replication in primary chick kidney (CK) cells and in chicken embryos was not affected. In chicken tracheal organ cultures (TOCs), the recombinant rIBVs reached titers similar to those of the wild-type virus, but in the case of viruses lacking the 3a protein, the titer declined reproducibly earlier. Translation of the IBV E protein is believed to be initiated by internal entry of ribosomes at a structure formed by the sequences corresponding to ORFs 3a and 3b. To assess the necessity of this mechanism, we deleted most of the sequence representing 3a and 3b to produce a gene in which ORF 3c (E) was adjacent to the gene 3 transcription-associated sequence. Western blot analysis revealed that the recombinant IBV produced fivefold less E protein. Nevertheless, titers produced in CK cells, embryos, and TOCs were similar to those of the wild-type virus, although they declined earlier in TOCs, probably due to the absence of the 3a protein. Thus, neither the tricistronic arrangement of gene 3, the internal initiation of translation of E protein, nor the 3a and 3b proteins are essential for replication per se, suggesting that these proteins are accessory proteins that may have roles in vivo.
Figures



Similar articles
-
Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus.J Virol. 1992 Oct;66(10):6143-54. doi: 10.1128/JVI.66.10.6143-6154.1992. J Virol. 1992. PMID: 1527853 Free PMC article.
-
A polycistronic mRNA specified by the coronavirus infectious bronchitis virus.Virology. 1991 Oct;184(2):531-44. doi: 10.1016/0042-6822(91)90423-9. Virology. 1991. PMID: 1653486 Free PMC article.
-
Identification of the avian infectious bronchitis coronaviruses with mutations in gene 3.Gene. 2008 Apr 15;412(1-2):12-25. doi: 10.1016/j.gene.2008.01.004. Epub 2008 Jan 19. Gene. 2008. PMID: 18295413 Free PMC article.
-
Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication.Virology. 2003 Jun 20;311(1):16-27. doi: 10.1016/s0042-6822(03)00117-x. Virology. 2003. PMID: 12832199 Free PMC article.
-
Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame.J Virol. 1996 Dec;70(12):8660-8. doi: 10.1128/JVI.70.12.8660-8668.1996. J Virol. 1996. PMID: 8970992 Free PMC article.
Cited by
-
Emerging infectious bronchitis virus (IBV) in Egypt: Evidence for an evolutionary advantage of a new S1 variant with a unique gene 3ab constellation.Infect Genet Evol. 2020 Nov;85:104433. doi: 10.1016/j.meegid.2020.104433. Epub 2020 Jul 1. Infect Genet Evol. 2020. PMID: 32622080 Free PMC article.
-
A DNA-based non-infectious replicon system to study SARS-CoV-2 RNA synthesis.Comput Struct Biotechnol J. 2022;20:5193-5202. doi: 10.1016/j.csbj.2022.08.044. Epub 2022 Aug 30. Comput Struct Biotechnol J. 2022. PMID: 36059866 Free PMC article.
-
Infectious Bronchitis Coronavirus Limits Interferon Production by Inducing a Host Shutoff That Requires Accessory Protein 5b.J Virol. 2016 Jul 27;90(16):7519-7528. doi: 10.1128/JVI.00627-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27279618 Free PMC article.
-
Accessory proteins of SARS-CoV and other coronaviruses.Antiviral Res. 2014 Sep;109:97-109. doi: 10.1016/j.antiviral.2014.06.013. Epub 2014 Jul 1. Antiviral Res. 2014. PMID: 24995382 Free PMC article. Review.
-
Genomic and single nucleotide polymorphism analysis of infectious bronchitis coronavirus.Infect Genet Evol. 2015 Jun;32:416-24. doi: 10.1016/j.meegid.2015.03.033. Epub 2015 Apr 3. Infect Genet Evol. 2015. PMID: 25843648 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
-
- Boursnell, M. E. G., T. D. K. Brown, I. J. Foulds, P. F. Green, F. M. Tomley, and M. M. Binns. 1987. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 68:57-77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

