Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2
- PMID: 16341264
- PMCID: PMC1307560
- DOI: 10.1172/JCI24787
Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2
Abstract
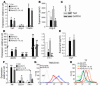
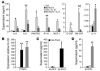
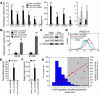
In the face of systemic risk factors, certain regions of the arterial vasculature remain relatively resistant to the development of atherosclerotic lesions. The biomechanically distinct environments in these arterial geometries exert a protective influence via certain key functions of the endothelial lining; however, the mechanisms underlying the coordinated regulation of specific mechano-activated transcriptional programs leading to distinct endothelial functional phenotypes have remained elusive. Here, we show that the transcription factor Kruppel-like factor 2 (KLF2) is selectively induced in endothelial cells exposed to a biomechanical stimulus characteristic of atheroprotected regions of the human carotid and that this flow-mediated increase in expression occurs via a MEK5/ERK5/MEF2 signaling pathway. Overexpression and silencing of KLF2 in the context of flow, combined with findings from genome-wide analyses of gene expression, demonstrate that the induction of KLF2 results in the orchestrated regulation of endothelial transcriptional programs controlling inflammation, thrombosis/hemostasis, vascular tone, and blood vessel development. Our data also indicate that KLF2 expression globally modulates IL-1beta-mediated endothelial activation. KLF2 therefore serves as a mechano-activated transcription factor important in the integration of multiple endothelial functions associated with regions of the arterial vasculature that are relatively resistant to atherogenesis.
Figures



Similar articles
-
Angiopoietin-1 induces Kruppel-like factor 2 expression through a phosphoinositide 3-kinase/AKT-dependent activation of myocyte enhancer factor 2.J Biol Chem. 2009 Feb 27;284(9):5592-601. doi: 10.1074/jbc.M806928200. Epub 2008 Dec 23. J Biol Chem. 2009. PMID: 19106103
-
Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells.Circulation. 2005 Aug 2;112(5):720-6. doi: 10.1161/CIRCULATIONAHA.104.525774. Epub 2005 Jul 25. Circulation. 2005. PMID: 16043642
-
KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation.J Exp Med. 2004 May 17;199(10):1305-15. doi: 10.1084/jem.20031132. Epub 2004 May 10. J Exp Med. 2004. PMID: 15136591 Free PMC article.
-
Transcription Factor KLF2 and Its Role in the Regulation of Inflammatory Processes.Biochemistry (Mosc). 2020 Jan;85(1):54-67. doi: 10.1134/S0006297920010058. Biochemistry (Mosc). 2020. PMID: 32079517 Review.
-
Key transcriptional regulators of the vasoprotective effects of shear stress.Hamostaseologie. 2009 Jan;29(1):39-40, 41-3. Hamostaseologie. 2009. PMID: 19151844 Review.
Cited by
-
A bypass flow model to study endothelial cell mechanotransduction across diverse flow environments.Mater Today Bio. 2024 Jun 13;27:101121. doi: 10.1016/j.mtbio.2024.101121. eCollection 2024 Aug. Mater Today Bio. 2024. PMID: 38988818 Free PMC article.
-
Transcriptional profile of the rat cardiovascular system at single cell resolution.bioRxiv [Preprint]. 2023 Nov 16:2023.11.14.567085. doi: 10.1101/2023.11.14.567085. bioRxiv. 2023. Update in: Cell Rep. 2024 Dec 21;44(1):115091. doi: 10.1016/j.celrep.2024.115091 PMID: 38014050 Free PMC article. Updated. Preprint.
-
Understanding the Causes and Implications of Endothelial Metabolic Variation in Cardiovascular Disease through Genome-Scale Metabolic Modeling.Front Cardiovasc Med. 2016 Apr 18;3:10. doi: 10.3389/fcvm.2016.00010. eCollection 2016. Front Cardiovasc Med. 2016. PMID: 27148541 Free PMC article. Review.
-
APOE-NOTCH axis governs elastogenesis during human cardiac valve remodeling.Nat Cardiovasc Res. 2024 Aug;3(8):933-950. doi: 10.1038/s44161-024-00510-3. Epub 2024 Jul 24. Nat Cardiovasc Res. 2024. PMID: 39196035
-
Venous thrombosis and obesity: from clinical needs to therapeutic challenges.Intern Emerg Med. 2024 Sep 13. doi: 10.1007/s11739-024-03765-7. Online ahead of print. Intern Emerg Med. 2024. PMID: 39269539 Review.
References
-
- Gimbrone MA, Jr, Topper JN, Nagel T, Anderson KR, Garcia-Cardena G. Endothelial dysfunction, hemodynamic forces, and atherogenesis. Ann. N. Y. Acad. Sci. 2000;902:230–239; discussion 239–240. - PubMed
-
- Traub O, Berk BC. Laminar shear stress: mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler. Thromb. Vasc. Biol. 1998;18:677–685. - PubMed
-
- Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc–dependent pathway. Nat. Immunol. 2001;2:698–704. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL072952/HL/NHLBI NIH HHS/United States
- R37-HL51150/HL/NHLBI NIH HHS/United States
- P50-HL56985/HL/NHLBI NIH HHS/United States
- R37 HL051150/HL/NHLBI NIH HHS/United States
- R01 HL076754/HL/NHLBI NIH HHS/United States
- R01 HL075427/HL/NHLBI NIH HHS/United States
- HL72952/HL/NHLBI NIH HHS/United States
- R01 HL069477/HL/NHLBI NIH HHS/United States
- P50 HL056985/HL/NHLBI NIH HHS/United States
- R01-HL076686/HL/NHLBI NIH HHS/United States
- HL-69477/HL/NHLBI NIH HHS/United States
- R01 HL076686/HL/NHLBI NIH HHS/United States
- HL75427/HL/NHLBI NIH HHS/United States
- HL-76754/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

