Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe
- PMID: 16341225
- PMCID: PMC1952686
- DOI: 10.1038/nsmb1033
Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe
Abstract
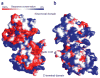
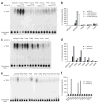
Decapping is a key step in both general and nonsense-mediated 5' --> 3' mRNA-decay pathways. Removal of the cap structure is catalyzed by the Dcp1-Dcp2 complex. The crystal structure of a C-terminally truncated Schizosaccharomyces pombe Dcp2p reveals two distinct domains: an all-helical N-terminal domain and a C-terminal domain that is a classic Nudix fold. The C-terminal domain of both Saccharomyces cerevisiae and S. pombe Dcp2p proteins is sufficient for decapping activity, although the N-terminal domain can affect the efficiency of Dcp2p function. The binding of Dcp2p to Dcp1p is mediated by a conserved surface on its N-terminal domain, and the N-terminal domain is required for Dcp1p to stimulate Dcp2p activity. The flexible nature of the N-terminal domain relative to the C-terminal domain suggests that Dcp1p binding to Dcp2p may regulate Dcp2p activity through conformational changes of the two domains.
Figures
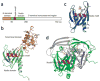
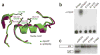


Similar articles
-
Structural basis of dcp2 recognition and activation by dcp1.Mol Cell. 2008 Feb 15;29(3):337-49. doi: 10.1016/j.molcel.2008.01.002. Mol Cell. 2008. PMID: 18280239 Free PMC article.
-
Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.Nat Struct Mol Biol. 2016 Jun;23(6):574-9. doi: 10.1038/nsmb.3232. Epub 2016 May 16. Nat Struct Mol Biol. 2016. PMID: 27183195
-
Structural basis of mRNA-cap recognition by Dcp1-Dcp2.Nat Struct Mol Biol. 2016 Nov;23(11):987-994. doi: 10.1038/nsmb.3301. Epub 2016 Oct 3. Nat Struct Mol Biol. 2016. PMID: 27694842 Free PMC article.
-
mRNA decapping: finding the right structures.Philos Trans R Soc Lond B Biol Sci. 2018 Nov 5;373(1762):20180164. doi: 10.1098/rstb.2018.0164. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 30397101 Free PMC article. Review.
-
New insights into the control of mRNA decapping.Trends Biochem Sci. 2006 May;31(5):241-3. doi: 10.1016/j.tibs.2006.03.001. Epub 2006 Mar 31. Trends Biochem Sci. 2006. PMID: 16580207 Review.
Cited by
-
Active site conformational dynamics are coupled to catalysis in the mRNA decapping enzyme Dcp2.Structure. 2013 Sep 3;21(9):1571-80. doi: 10.1016/j.str.2013.06.021. Epub 2013 Aug 1. Structure. 2013. PMID: 23911090 Free PMC article.
-
Length-dependent prediction of protein intrinsic disorder.BMC Bioinformatics. 2006 Apr 17;7:208. doi: 10.1186/1471-2105-7-208. BMC Bioinformatics. 2006. PMID: 16618368 Free PMC article.
-
Involvement of fission yeast Pdc2 in RNA degradation and P-body function.RNA. 2017 Apr;23(4):493-503. doi: 10.1261/rna.059766.116. Epub 2016 Dec 28. RNA. 2017. PMID: 28031482 Free PMC article.
-
Cap analogs modified with 1,2-dithiodiphosphate moiety protect mRNA from decapping and enhance its translational potential.Nucleic Acids Res. 2016 Nov 16;44(20):9578-9590. doi: 10.1093/nar/gkw896. Epub 2016 Oct 7. Nucleic Acids Res. 2016. PMID: 27903882 Free PMC article.
-
The Activity-Dependent Regulation of Protein Kinase Stability by the Localization to P-Bodies.Genetics. 2016 Jul;203(3):1191-202. doi: 10.1534/genetics.116.187419. Epub 2016 May 6. Genetics. 2016. PMID: 27182950 Free PMC article.
References
-
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. - PubMed
-
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. - PubMed
-
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

