Single-site oxidation, cysteine 108 to cysteine sulfinic acid, in D-amino acid oxidase from Trigonopsis variabilis and its structural and functional consequences
- PMID: 16332786
- PMCID: PMC1317377
- DOI: 10.1128/AEM.71.12.8061-8068.2005
Single-site oxidation, cysteine 108 to cysteine sulfinic acid, in D-amino acid oxidase from Trigonopsis variabilis and its structural and functional consequences
Abstract
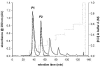
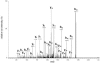
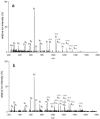
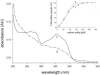
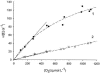
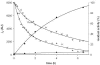
One of the primary sources of enzyme instability is protein oxidative modification triggering activity loss or denaturation. We show here that the side chain of Cys108 is the main site undergoing stress-induced oxidation in Trigonopsis variabilis d-amino acid oxidase, a flavoenzyme employed industrially for the conversion of cephalosporin C. High-resolution anion-exchange chromatography was used to separate the reduced and oxidized protein forms, which constitute, in a molar ratio of about 3:1, the active biocatalyst isolated from the yeast. Comparative analysis of their tryptic peptides by electrospray tandem mass spectrometry allowed unequivocal assignment of the modification as the oxidation of Cys108 into cysteine sulfinic acid. Cys108 is likely located on a surface-exposed protein region within the flavin adenine dinucleotide (FAD) binding domain, but remote from the active center. Its oxidized side chain was remarkably stable in solution, thus enabling the relative biochemical characterization of native and modified enzyme forms. The oxidation of Cys108 causes a global conformational response that affects the protein environment of the FAD cofactor. In comparison with the native enzyme, it results in a fourfold-decreased specific activity, reflecting a catalytic efficiency for reduction of dioxygen lowered by about the same factor, and a markedly decreased propensity to aggregate under conditions of thermal denaturation. These results open up unprecedented routes for stabilization of the oxidase and underscore the possible significance of protein chemical heterogeneity for biocatalyst function and stability.
Figures
Similar articles
-
The role of Cys108 in Trigonopsis variabilis d-amino acid oxidase examined through chemical oxidation studies and point mutations C108S and C108D.Biochim Biophys Acta. 2010 Jul;1804(7):1483-91. doi: 10.1016/j.bbapap.2010.02.009. Epub 2010 Mar 1. Biochim Biophys Acta. 2010. PMID: 20193780
-
Stability and stabilization of D-amino acid oxidase from the yeast Trigonopsis variabilis.Biochem Soc Trans. 2007 Dec;35(Pt 6):1588-92. doi: 10.1042/BST0351588. Biochem Soc Trans. 2007. PMID: 18031272 Review.
-
Thermal inactivation of D-amino acid oxidase from Trigonopsis variabilis occurs via three parallel paths of irreversible denaturation.Biotechnol Bioeng. 2006 Jul 5;94(4):645-54. doi: 10.1002/bit.20854. Biotechnol Bioeng. 2006. PMID: 16538681
-
The role of cofactor binding in tryptophan accessibility and conformational stability of His-tagged D-amino acid oxidase from Trigonopsis variabilis.Biochim Biophys Acta. 2007 May;1774(5):556-65. doi: 10.1016/j.bbapap.2007.03.009. Epub 2007 Mar 24. Biochim Biophys Acta. 2007. PMID: 17466607
-
D-Amino acid oxidase: structure, catalytic mechanism, and practical application.Biochemistry (Mosc). 2005 Jan;70(1):40-54. Biochemistry (Mosc). 2005. PMID: 15701048 Review.
Cited by
-
Multipoint TvDAAO Mutants for Cephalosporin C Bioconversion.Int J Mol Sci. 2019 Sep 7;20(18):4412. doi: 10.3390/ijms20184412. Int J Mol Sci. 2019. PMID: 31500317 Free PMC article.
-
Thermal Inactivation of a Cold-Active Esterase PMGL3 Isolated from the Permafrost Metagenomic Library.Biomolecules. 2019 Dec 16;9(12):880. doi: 10.3390/biom9120880. Biomolecules. 2019. PMID: 31888238 Free PMC article.
-
Cofactor binding protects flavodoxin against oxidative stress.PLoS One. 2012;7(7):e41363. doi: 10.1371/journal.pone.0041363. Epub 2012 Jul 19. PLoS One. 2012. PMID: 22829943 Free PMC article.
-
The redox biochemistry of protein sulfenylation and sulfinylation.J Biol Chem. 2013 Sep 13;288(37):26480-8. doi: 10.1074/jbc.R113.467738. Epub 2013 Jul 16. J Biol Chem. 2013. PMID: 23861405 Free PMC article. Review.
-
A novel thermostable D-amino acid oxidase of the thermophilic fungus Rasamsonia emersonii strain YA.Sci Rep. 2019 Aug 16;9(1):11948. doi: 10.1038/s41598-019-48480-y. Sci Rep. 2019. PMID: 31420577 Free PMC article.
References
-
- Alaejos, M. S., and F. J. G. Montelongo. 2004. Application of amperometric biosensors to the determination of vitamins and α-amino acids. Chem. Rev. 104:3239-3265. - PubMed
-
- Betancor, L., A. Hidalgo, G. Fernandez-Lorente, C. Mateo, V. Rodriguez, M. Fuentes, F. Lopez-Gallego, R. Fernandez-Lafuente, and J. M. Guisan. 2003. Use of physicochemical tools to determine the choice of optimal enzyme: stabilization of d-amino acid oxidase. Biotechnol. Prog. 19:784-788. - PubMed
-
- Biteau, B., J. Labarre, and M. B. Toledano. 2003. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425:980-984. - PubMed
-
- Burton, S. G. 2003. Oxidizing enzymes as biocatalysts. Trends Biotechnol. 21:543-549. - PubMed
-
- Choi, M. H., I. K. Lee, G. W. Kim, B. U. Kim, Y.- H. Han, D.-Y. Yu, H. S. Park, K. Y. Kim, J. S. Lee, C. Choi, Y. S. Bae, B. I. Lee, S. G. Rhee, and S. W. Kang. 2005. Regulation of PDGF signalling and vascular remodeling by peroxiredoxin II. Nature 435:347-353. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

