Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin
- PMID: 16330710
- PMCID: PMC2171291
- DOI: 10.1083/jcb.200506083
Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin
Abstract
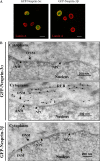
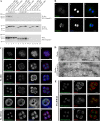
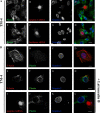
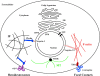
Despite their importance in cell biology, the mechanisms that maintain the nucleus in its proper position in the cell are not well understood. This is primarily the result of an incomplete knowledge of the proteins in the outer nuclear membrane (ONM) that are able to associate with the different cytoskeletal systems. Two related ONM proteins, nuclear envelope spectrin repeat (nesprin)-1 and -2, are known to make direct connections with the actin cytoskeleton through their NH2-terminal actin-binding domain (ABD). We have now isolated a third member of the nesprin family that lacks an ABD and instead binds to the plakin family member plectin, which can associate with the intermediate filament (IF) system. Overexpression of nesprin-3 results in a dramatic recruitment of plectin to the nuclear perimeter, which is where these two molecules are colocalized with both keratin-6 and -14. Importantly, plectin binds to the integrin alpha6beta4 at the cell surface and to nesprin-3 at the ONM in keratinocytes, suggesting that there is a continuous connection between the nucleus and the extracellular matrix through the IF cytoskeleton.
Figures



Similar articles
-
Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin.J Cell Sci. 2007 Oct 1;120(Pt 19):3384-94. doi: 10.1242/jcs.014191. J Cell Sci. 2007. PMID: 17881500
-
Nesprin-3 augments peripheral nuclear localization of intermediate filaments in zebrafish.J Cell Sci. 2011 Mar 1;124(Pt 5):755-64. doi: 10.1242/jcs.081174. Epub 2011 Feb 8. J Cell Sci. 2011. PMID: 21303928
-
Nesprin-3 connects plectin and vimentin to the nuclear envelope of Sertoli cells but is not required for Sertoli cell function in spermatogenesis.Mol Biol Cell. 2013 Aug;24(15):2454-66. doi: 10.1091/mbc.E13-02-0100. Epub 2013 Jun 12. Mol Biol Cell. 2013. PMID: 23761073 Free PMC article.
-
Nesprin-3: a versatile connector between the nucleus and the cytoskeleton.Biochem Soc Trans. 2011 Dec;39(6):1719-24. doi: 10.1042/BST20110669. Biochem Soc Trans. 2011. PMID: 22103514 Review.
-
A Glance at the Nuclear Envelope Spectrin Repeat Protein 3.Biomed Res Int. 2019 Nov 20;2019:1651805. doi: 10.1155/2019/1651805. eCollection 2019. Biomed Res Int. 2019. PMID: 31828088 Free PMC article. Review.
Cited by
-
Keeping the LINC: the importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function.Biochem Soc Trans. 2011 Dec;39(6):1729-34. doi: 10.1042/BST20110686. Biochem Soc Trans. 2011. PMID: 22103516 Free PMC article. Review.
-
Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling.Hum Mol Genet. 2013 Jun 15;22(12):2335-49. doi: 10.1093/hmg/ddt079. Epub 2013 Feb 19. Hum Mol Genet. 2013. PMID: 23427149 Free PMC article.
-
The expanding significance of keratin intermediate filaments in normal and diseased epithelia.Curr Opin Cell Biol. 2013 Feb;25(1):47-56. doi: 10.1016/j.ceb.2012.10.018. Epub 2012 Dec 25. Curr Opin Cell Biol. 2013. PMID: 23270662 Free PMC article. Review.
-
Properties of cells through life and death - an acoustic microscopy investigation.Cell Cycle. 2015;14(18):2891-8. doi: 10.1080/15384101.2015.1069925. Cell Cycle. 2015. PMID: 26178635 Free PMC article.
-
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.J Biol Chem. 2016 Sep 2;291(36):18643-62. doi: 10.1074/jbc.M116.732909. Epub 2016 Jul 13. J Biol Chem. 2016. PMID: 27413182 Free PMC article.
References
-
- Apel, E.D., R.M. Lewis, R.M. Grady, and J.R. Sanes. 2000. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275:31986–31995. - PubMed
-
- Borradori, L., and A. Sonnenberg. 1999. Structure and function of hemidesmosomes: more than simple adhesion complexes. J. Invest. Dermatol. 112:411–418. - PubMed
-
- Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553. - PubMed
-
- Burke, B., and C.L. Stewart. 2002. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 3:575–585. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

