Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells
- PMID: 16322558
- PMCID: PMC1315390
- DOI: 10.1101/gad.1369805
Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells
Abstract
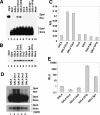
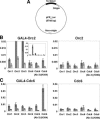
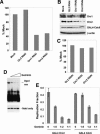
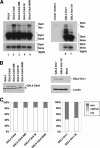
Origins of replication are expected to recruit initiation proteins like origin recognition complex (ORC) and Cdc6 in eukaryotes and provide a platform for unwinding DNA. Here we test whether localization of initiation proteins onto DNA is sufficient for origin function. Different components of the ORC complex and Cdc6 stimulated prereplicative complex (pre-RC) formation and replication initiation when fused to the GAL4 DNA-binding domain and recruited to plasmid DNA containing a tandem array of GAL4-binding sites. Replication occurred once per cell cycle and was inhibited by Geminin, indicating that the plasmid was properly licensed during the cell cycle. The GAL4 fusion protein recruits other polypeptides of the ORC-Cdc6 complex, and nascent strand abundance was highest near the GAL4-binding sites. Therefore, the artificial origin recapitulates many of the regulatory features of physiological origins and is valuable for studies on replication initiation in mammalian cells. We demonstrated the utility of this system by showing the functional importance of the ATPase domains of human Cdc6 and Orc1 and the dispensability of the N-terminal segments of Orc1 and Orc2 in this assay. Artificial recruitment of a eukaryotic cellular replication initiation factor to a DNA sequence can create a functional origin of replication, providing a robust genetic assay for these factors and a novel approach to generating episomal vectors for gene therapy.
Figures

Similar articles
-
Epstein-Barr nuclear antigen 1 (EBNA1)-dependent recruitment of origin recognition complex (Orc) on oriP of Epstein-Barr virus with purified proteins: stimulation by Cdc6 through its direct interaction with EBNA1.J Biol Chem. 2012 Jul 6;287(28):23977-94. doi: 10.1074/jbc.M112.368456. Epub 2012 May 14. J Biol Chem. 2012. PMID: 22589552 Free PMC article.
-
Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase.Mol Cell. 2006 Jan 6;21(1):29-39. doi: 10.1016/j.molcel.2005.11.023. Mol Cell. 2006. PMID: 16387651
-
The structure of ORC-Cdc6 on an origin DNA reveals the mechanism of ORC activation by the replication initiator Cdc6.Nat Commun. 2021 Jun 23;12(1):3883. doi: 10.1038/s41467-021-24199-1. Nat Commun. 2021. PMID: 34162887 Free PMC article.
-
DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity.Biochem Cell Biol. 2010 Feb;88(1):49-62. doi: 10.1139/o09-154. Biochem Cell Biol. 2010. PMID: 20130679 Review.
-
Control of DNA replication licensing in a cell cycle.Genes Cells. 2002 Jun;7(6):523-34. doi: 10.1046/j.1365-2443.2002.00544.x. Genes Cells. 2002. PMID: 12059957 Review.
Cited by
-
Epstein-Barr nuclear antigen 1 (EBNA1)-dependent recruitment of origin recognition complex (Orc) on oriP of Epstein-Barr virus with purified proteins: stimulation by Cdc6 through its direct interaction with EBNA1.J Biol Chem. 2012 Jul 6;287(28):23977-94. doi: 10.1074/jbc.M112.368456. Epub 2012 May 14. J Biol Chem. 2012. PMID: 22589552 Free PMC article.
-
Initiation of replication in Xenopus egg extracts at a spatially defined origin.Cell Cycle. 2015 Aug 3;14(15):2391. doi: 10.1080/15384101.2015.1063306. Epub 2015 Jun 19. Cell Cycle. 2015. PMID: 26091433 Free PMC article. No abstract available.
-
Regulation of the initiation of DNA replication in human cells.DNA Repair (Amst). 2018 Dec;72:99-106. doi: 10.1016/j.dnarep.2018.09.003. Epub 2018 Sep 12. DNA Repair (Amst). 2018. PMID: 30266203 Free PMC article. Review.
-
A dual role for the chromatin reader ORCA/LRWD1 in targeting the origin recognition complex to chromatin.EMBO J. 2023 Sep 18;42(18):e114654. doi: 10.15252/embj.2023114654. Epub 2023 Aug 8. EMBO J. 2023. PMID: 37551430 Free PMC article.
-
REV1 restrains DNA polymerase zeta to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo.Nucleic Acids Res. 2008 Dec;36(21):6767-80. doi: 10.1093/nar/gkn651. Epub 2008 Oct 25. Nucleic Acids Res. 2008. PMID: 18953031 Free PMC article.
References
-
- Aggarwal, B.D. and Calvi, B.R. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372-376. - PubMed
-
- Aladjem, M.I., Rodewald, L.W., Lin, C.M., Bowman, S., Cimbora, D.M., Brody, L.L., Epner, E.M., Groudine, M., and Wahl, G.M. 2002. Replication initiation patterns in the β-globin loci of totipotent and differentiated murine cells: Evidence for multiple initiation regions. Mol. Cell. Biol. 22: 442-452. - PMC - PubMed
-
- Bell, S.P. and Dutta, A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333-374. - PubMed
-
- Bell, S.P. and Stillman, B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128-134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
