P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes
- PMID: 16308568
- PMCID: PMC1356315
- DOI: 10.1038/sj.emboj.7600876
P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes
Abstract
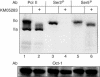
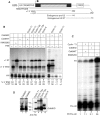
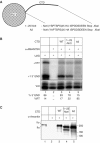
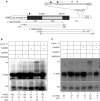
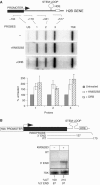
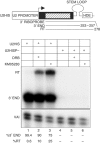
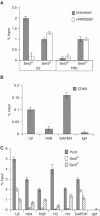
Phosphorylation of Ser2 of the heptapeptide repeat of the CTD of mammalian pol II by P-TEFb is associated with productive elongation of transcription of protein-coding genes. Here, we show that the CTD of pol II transcribing the human U2 snRNA genes is phosphorylated on Ser2 in vivo and that both the CDK9 kinase and cyclin T components of P-TEFb are required for cotranscriptional recognition of the 3' box RNA 3' end processing signal. However, inhibitors of CDK9 do not affect transcription of the U2 genes, indicating that P-TEFb functions exclusively as an RNA processing factor in expression of these relatively short, intronless genes. We also show that inhibition of CDK9 does not adversely affect either transcription of an intron-less, replication-activated histone H2b gene or recognition of the histone gene-specific U7-dependent RNA 3' end formation signal. These results emphasize that the role of P-TEFb as an activator of transcription elongation can be separated from its role in RNA processing and that neither function is universally required for expression of mammalian pol II-dependent genes.
Figures

Similar articles
-
Chromatin structure is implicated in "late" elongation checkpoints on the U2 snRNA and beta-actin genes.Mol Cell Biol. 2009 Jul;29(14):4002-13. doi: 10.1128/MCB.00189-09. Epub 2009 May 18. Mol Cell Biol. 2009. PMID: 19451231 Free PMC article.
-
7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes.Nature. 2001 Nov 15;414(6861):322-5. doi: 10.1038/35104581. Nature. 2001. PMID: 11713533
-
A positive feedback loop links opposing functions of P-TEFb/Cdk9 and histone H2B ubiquitylation to regulate transcript elongation in fission yeast.PLoS Genet. 2012;8(8):e1002822. doi: 10.1371/journal.pgen.1002822. Epub 2012 Aug 2. PLoS Genet. 2012. PMID: 22876190 Free PMC article.
-
Expression of human snRNA genes from beginning to end.Biochem Soc Trans. 2008 Aug;36(Pt 4):590-4. doi: 10.1042/BST0360590. Biochem Soc Trans. 2008. PMID: 18631122 Review.
-
P-TEFb goes viral.Bioessays. 2016 Jul;38 Suppl 1:S75-85. doi: 10.1002/bies.201670912. Bioessays. 2016. PMID: 27417125 Review.
Cited by
-
Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes.Mol Cell. 2012 Jan 13;45(1):111-22. doi: 10.1016/j.molcel.2011.11.006. Epub 2011 Dec 1. Mol Cell. 2012. PMID: 22137580 Free PMC article.
-
A novel TBP-TAF complex on RNA polymerase II-transcribed snRNA genes.Transcription. 2012 Mar-Apr;3(2):92-104. doi: 10.4161/trns.19783. Epub 2012 Mar 1. Transcription. 2012. PMID: 22441827 Free PMC article.
-
Complex effects of flavopiridol on the expression of primary response genes.Cell Div. 2012 Mar 29;7:11. doi: 10.1186/1747-1028-7-11. Cell Div. 2012. PMID: 22458775 Free PMC article.
-
MLL-AF9 and MLL-ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia.Exp Hematol. 2011 Jan;39(1):77-86.e1-5. doi: 10.1016/j.exphem.2010.09.003. Epub 2010 Sep 18. Exp Hematol. 2011. PMID: 20854876 Free PMC article.
-
Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II.J Virol. 2007 May;81(10):5091-101. doi: 10.1128/JVI.00184-07. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344289 Free PMC article.
References
-
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 - PubMed
-
- Chao SH, Price DH (2001) Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276: 31793–31799 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

