Phylogenetic incongruence among oncogenic genital alpha human papillomaviruses
- PMID: 16306621
- PMCID: PMC1316001
- DOI: 10.1128/JVI.79.24.15503-15510.2005
Phylogenetic incongruence among oncogenic genital alpha human papillomaviruses
Abstract
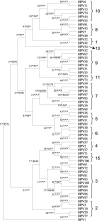
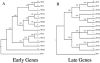
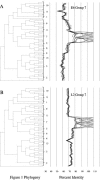
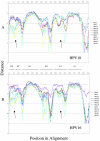
The human papillomaviruses (HPVs) have long been thought to follow a monophyletic pattern of evolution with little if any evidence for recombination between genomes. On the basis of this model, both oncogenicity and tissue tropism appear to have evolved once. Still, no systematic statistical analyses have shown whether monophyly is the rule across all HPV open reading frames (ORFs). We conducted a taxonomic analysis of 59 mucosal/genital HPVs using whole-genome and sliding-window similarity measures; maximum-parsimony, neighbor-joining, and Bayesian phylogenetic analyses; and localized incongruence length difference (LILD) analyses. The algorithm for the LILD analyses localized incongruence by calculating the tree length differences between constrained and unconstrained nodes in a total-evidence tree across all HPV ORFs. The process allows statistical evaluation of every ORF/node pair in the total-evidence tree. The most significant incongruence was observed at the putative high-risk (i.e., cancer-associated) node, the common oncogenic ancestor for alpha HPV species 9 (e.g., HPV type 16 [HPV16]), 11, 7 (e.g., HPV18), 5, and 6. Although these groups share early-gene homology, including high degrees of similarity among E6 and E7, groups 9 and 11 diverge from groups 7, 5, and 6 with respect to L2 and L1. The HPV species groups primarily associated with cervical and anogenital cancers appear to follow two distinct evolutionary paths, one conferred by the early genes and another by the late genes. The incongruence in the genital HPV phylogeny could have occurred from an early recombination event, an ecological niche change, and/or asymmetric genome convergence driven by intense selection. These data indicate that the phylogeny of the oncogenic HPVs is complex and that their evolution may not be monophyletic across all genes.
Figures
Similar articles
-
Biology and pathological associations of the human papillomaviruses: a review.Malays J Pathol. 1998 Jun;20(1):1-10. Malays J Pathol. 1998. PMID: 10879257 Review.
-
Human papillomavirus (HPV) types 101 and 103 isolated from cervicovaginal cells lack an E6 open reading frame (ORF) and are related to gamma-papillomaviruses.Virology. 2007 Apr 10;360(2):447-53. doi: 10.1016/j.virol.2006.10.022. Epub 2006 Nov 22. Virology. 2007. PMID: 17125811 Free PMC article.
-
Genetic diversity of human papillomavirus type 16 E6, E7, and L1 genes in Italian women with different grades of cervical lesions.J Med Virol. 2009 Sep;81(9):1627-34. doi: 10.1002/jmv.21552. J Med Virol. 2009. PMID: 19626616
-
Human papillomaviruses: genetic basis of carcinogenicity.Public Health Genomics. 2009;12(5-6):281-90. doi: 10.1159/000214919. Epub 2009 Aug 11. Public Health Genomics. 2009. PMID: 19684441 Free PMC article. Review.
-
Phylogenetic analysis of beta-papillomaviruses as inferred from nucleotide and amino acid sequence data.Mol Phylogenet Evol. 2007 Jan;42(1):213-22. doi: 10.1016/j.ympev.2006.07.011. Epub 2006 Jul 26. Mol Phylogenet Evol. 2007. PMID: 16935527
Cited by
-
Genomic diversity and interspecies host infection of alpha12 Macaca fascicularis papillomaviruses (MfPVs).Virology. 2009 Oct 25;393(2):304-10. doi: 10.1016/j.virol.2009.07.012. Epub 2009 Aug 28. Virology. 2009. PMID: 19716580 Free PMC article.
-
Genome Plasticity in Papillomaviruses and De Novo Emergence of E5 Oncogenes.Genome Biol Evol. 2019 Jun 1;11(6):1602-1617. doi: 10.1093/gbe/evz095. Genome Biol Evol. 2019. PMID: 31076746 Free PMC article.
-
K-Mer Analyses Reveal Different Evolutionary Histories of Alpha, Beta, and Gamma Papillomaviruses.Int J Mol Sci. 2021 Sep 6;22(17):9657. doi: 10.3390/ijms22179657. Int J Mol Sci. 2021. PMID: 34502564 Free PMC article.
-
Isolation of three novel rat and mouse papillomaviruses and their genomic characterization.PLoS One. 2012;7(10):e47164. doi: 10.1371/journal.pone.0047164. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077564 Free PMC article.
-
Papillomaviruses infecting cetaceans exhibit signs of genome adaptation following a recombination event.Virus Evol. 2020 Jun 30;6(1):veaa038. doi: 10.1093/ve/veaa038. eCollection 2020 Jan. Virus Evol. 2020. PMID: 32665861 Free PMC article.
References
-
- Agosti, D., D. Jacobs, and R. DeSalle. 1996. On combining protein sequences and nucleic acid sequences in phylogenetic analysis: the homeobox protein case. Cladistics 12:65-82. - PubMed
-
- Burk, R. D., M. Terai, P. E. Gravitt, L. A. Brinton, R. J. Kurman, W. A. Barnes, M. D. Greenberg, O. C. Hadjimichael, L. Fu, L. McGowan, R. Mortel, P. E. Schwartz, and A. Hildesheim. 2003. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer Res. 63:7215-7220. - PubMed
-
- de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. Zur Hausen. 2004. Classification of papillomaviruses. Virology 324:17-27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

