Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi
- PMID: 16282589
- PMCID: PMC1283529
- DOI: 10.1093/nar/gki952
Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi
Abstract
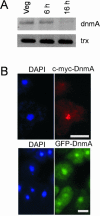
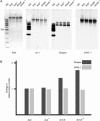
We have identified a DNA methyltransferase of the Dnmt2 family in Dictyostelium that was denominated DnmA. Expression of the dnmA gene is downregulated during the developmental cycle. Overall DNA methylation in Dictyostelium is approximately 0.2% of the cytosine residues, which indicates its restriction to a limited set of genomic loci. Bisulfite sequencing of specific sites revealed that DnmA is responsible for methylation of mostly asymmetric C-residues in the retrotransposons DIRS-1 and Skipper. Disruption of the gene resulted in a loss of methylation and in increased transcription and mobilization of Skipper. Skipper transcription was also upregulated in strains that had genes encoding components of the RNA interference pathway disrupted. In contrast, DIRS-1 expression was not affected by a loss of DnmA but was strongly increased in strains that had the RNA-directed RNA polymerase gene rrpC disrupted. A large number of siRNAs were found that corresponded to the DIRS-1 sequence, suggesting concerted regulation of DIRS-1 expression by RNAi and DNA modification. No siRNAs corresponding to the standard Skipper element were found. The data show that DNA methylation plays a crucial role in epigenetic gene silencing in Dictyostelium but that different, partially overlapping mechanisms control transposon silencing.
Figures

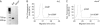





Similar articles
-
Developmentally regulated DNA methylation in Dictyostelium discoideum.Eukaryot Cell. 2006 Jan;5(1):18-25. doi: 10.1128/EC.5.1.18-25.2006. Eukaryot Cell. 2006. PMID: 16400165 Free PMC article.
-
Argonaute proteins affect siRNA levels and accumulation of a novel extrachromosomal DNA from the Dictyostelium retrotransposon DIRS-1.J Biol Chem. 2014 Dec 19;289(51):35124-38. doi: 10.1074/jbc.M114.612663. Epub 2014 Oct 28. J Biol Chem. 2014. PMID: 25352599 Free PMC article.
-
The Dictyostelium discoideum RNA-dependent RNA polymerase RrpC silences the centromeric retrotransposon DIRS-1 post-transcriptionally and is required for the spreading of RNA silencing signals.Nucleic Acids Res. 2014 Mar;42(5):3330-45. doi: 10.1093/nar/gkt1337. Epub 2013 Dec 24. Nucleic Acids Res. 2014. PMID: 24369430 Free PMC article.
-
DIRS-1 and the other tyrosine recombinase retrotransposons.Cytogenet Genome Res. 2005;110(1-4):575-88. doi: 10.1159/000084991. Cytogenet Genome Res. 2005. PMID: 16093711 Review.
-
Epigenetic decisions in mammalian germ cells.Science. 2007 Apr 20;316(5823):398-9. doi: 10.1126/science.1137544. Science. 2007. PMID: 17446388 Review.
Cited by
-
Global characterization of the Dicer-like protein DrnB roles in miRNA biogenesis in the social amoeba Dictyostelium discoideum.RNA Biol. 2018;15(7):937-954. doi: 10.1080/15476286.2018.1481697. Epub 2018 Aug 21. RNA Biol. 2018. Update in: RNA Biol. 2018;15(7):iii. doi: 10.1080/15476286.2018.1525144. PMID: 29966484 Free PMC article. Updated.
-
De novo search for non-coding RNA genes in the AT-rich genome of Dictyostelium discoideum: performance of Markov-dependent genome feature scoring.Genome Res. 2008 Jun;18(6):888-99. doi: 10.1101/gr.069104.107. Epub 2008 Mar 17. Genome Res. 2008. PMID: 18347326 Free PMC article.
-
The Biomphalaria glabrata DNA methylation machinery displays spatial tissue expression, is differentially active in distinct snail populations and is modulated by interactions with Schistosoma mansoni.PLoS Negl Trop Dis. 2017 May 16;11(5):e0005246. doi: 10.1371/journal.pntd.0005246. eCollection 2017 May. PLoS Negl Trop Dis. 2017. PMID: 28510608 Free PMC article.
-
Differential effects of heterochromatin protein 1 isoforms on mitotic chromosome distribution and growth in Dictyostelium discoideum.Eukaryot Cell. 2006 Mar;5(3):530-43. doi: 10.1128/EC.5.3.530-543.2006. Eukaryot Cell. 2006. PMID: 16524908 Free PMC article.
-
Cytosine Methylation Within Marine Sediment Microbial Communities: Potential Epigenetic Adaptation to the Environment.Front Microbiol. 2019 Jun 11;10:1291. doi: 10.3389/fmicb.2019.01291. eCollection 2019. Front Microbiol. 2019. PMID: 31244806 Free PMC article.
References
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature Rev. Genet. 2002;3:662–673. - PubMed
-
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–622. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

