The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure
- PMID: 16214138
- PMCID: PMC7094587
- DOI: 10.1016/j.febslet.2005.09.038
The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure
Abstract
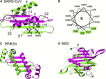
We have employed NMR to investigate the structure of SARS coronavirus nucleocapsid protein dimer. We found that the secondary structure of the dimerization domain consists of five alpha helices and a beta-hairpin. The dimer interface consists of a continuous four-stranded beta-sheet superposed by two long alpha helices, reminiscent of that found in the nucleocapsid protein of porcine respiratory and reproductive syndrome virus. Extensive hydrogen bond formation between the two hairpins and hydrophobic interactions between the beta-sheet and the alpha helices render the interface highly stable. Sequence alignment suggests that other coronavirus may share the same structural topology.
Figures



Similar articles
-
Structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus.Structure. 2003 Nov;11(11):1445-51. doi: 10.1016/j.str.2003.09.018. Structure. 2003. PMID: 14604534 Free PMC article.
-
Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method.J Mol Biol. 2008 Jul 18;380(4):608-22. doi: 10.1016/j.jmb.2007.11.093. Epub 2007 Dec 5. J Mol Biol. 2008. PMID: 18561946 Free PMC article.
-
Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae.J Biol Chem. 2006 Jun 23;281(25):17134-17139. doi: 10.1074/jbc.M602107200. Epub 2006 Apr 20. J Biol Chem. 2006. PMID: 16627473 Free PMC article.
-
The SARS coronavirus nucleocapsid protein--forms and functions.Antiviral Res. 2014 Mar;103:39-50. doi: 10.1016/j.antiviral.2013.12.009. Epub 2014 Jan 11. Antiviral Res. 2014. PMID: 24418573 Free PMC article. Review.
-
The SARS-CoV nucleocapsid protein: a protein with multifarious activities.Infect Genet Evol. 2008 Jul;8(4):397-405. doi: 10.1016/j.meegid.2007.07.004. Epub 2007 Jul 20. Infect Genet Evol. 2008. PMID: 17881296 Free PMC article. Review.
Cited by
-
The SARS-CoV-2 nucleoprotein associates with anionic lipid membranes.J Biol Chem. 2024 Aug;300(8):107456. doi: 10.1016/j.jbc.2024.107456. Epub 2024 Jun 10. J Biol Chem. 2024. PMID: 38866325 Free PMC article.
-
Peptide-based inhibitors hold great promise as the broad-spectrum agents against coronavirus.Front Microbiol. 2023 Jan 19;13:1093646. doi: 10.3389/fmicb.2022.1093646. eCollection 2022. Front Microbiol. 2023. PMID: 36741878 Free PMC article. Review.
-
Targeting the N-Terminus Domain of the Coronavirus Nucleocapsid Protein Induces Abnormal Oligomerization via Allosteric Modulation.Front Mol Biosci. 2022 Apr 19;9:871499. doi: 10.3389/fmolb.2022.871499. eCollection 2022. Front Mol Biosci. 2022. PMID: 35517857 Free PMC article.
-
Structural and conformational analysis of SARS CoV 2 N-CTD revealing monomeric and dimeric active sites during the RNA-binding and stabilization: Insights towards potential inhibitors for N-CTD.Comput Biol Med. 2021 Jul;134:104495. doi: 10.1016/j.compbiomed.2021.104495. Epub 2021 May 15. Comput Biol Med. 2021. PMID: 34022485 Free PMC article.
-
The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein.Nat Commun. 2021 Jan 21;12(1):502. doi: 10.1038/s41467-020-20768-y. Nat Commun. 2021. PMID: 33479198 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

