In vivo functional characterization of the SARS-Coronavirus 3a protein in Drosophila
- PMID: 16212942
- PMCID: PMC7117541
- DOI: 10.1016/j.bbrc.2005.09.098
In vivo functional characterization of the SARS-Coronavirus 3a protein in Drosophila
Abstract
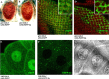
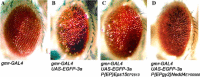
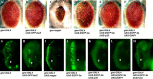
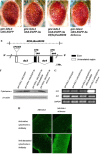
The Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3a locus encodes a 274 a.a. novel protein, and its expression has been confirmed in SARS patients. To study functional roles of 3a, we established a transgenic fly model for the SARS-CoV 3a gene. Misexpression of 3a in Drosophila caused a dominant rough eye phenotype. Using a specific monoclonal antibody, we demonstrated that the 3a protein displayed a punctate cytoplasmic localization in Drosophila as in SARS-CoV-infected cells. We provide genetic evidence to support that 3a is functionally related to clathrin-mediated endocytosis. We further found that 3a misexpression induces apoptosis, which could be modulated by cellular cytochrome c levels and caspase activity. From a forward genetic screen, 78 dominant 3a modifying loci were recovered and the identity of these modifiers revealed that the severity of the 3a-induced rough eye phenotype depends on multiple cellular processes including gene transcriptional regulation.
Figures

Similar articles
-
Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis.mBio. 2018 May 22;9(3):e02325-17. doi: 10.1128/mBio.02325-17. mBio. 2018. PMID: 29789363 Free PMC article.
-
Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients.J Mol Biol. 2004 Jul 30;341(1):271-9. doi: 10.1016/j.jmb.2004.06.016. J Mol Biol. 2004. PMID: 15312778 Free PMC article.
-
Genetic lesions within the 3a gene of SARS-CoV.Virol J. 2005 Jun 20;2:51. doi: 10.1186/1743-422X-2-51. Virol J. 2005. PMID: 15963240 Free PMC article.
-
Expression of the severe acute respiratory syndrome coronavirus 3a protein and the assembly of coronavirus-like particles in the baculovirus expression system.Methods Mol Biol. 2007;379:35-50. doi: 10.1007/978-1-59745-393-6_3. Methods Mol Biol. 2007. PMID: 17502669 Free PMC article. Review.
-
Coronavirus Proteins as Ion Channels: Current and Potential Research.Front Immunol. 2020 Oct 9;11:573339. doi: 10.3389/fimmu.2020.573339. eCollection 2020. Front Immunol. 2020. PMID: 33154751 Free PMC article. Review.
Cited by
-
A comprehensive Drosophila resource to identify key functional interactions between SARS-CoV-2 factors and host proteins.Cell Rep. 2023 Aug 29;42(8):112842. doi: 10.1016/j.celrep.2023.112842. Epub 2023 Jul 20. Cell Rep. 2023. PMID: 37480566 Free PMC article.
-
A Drosophila model of HPV16-induced cancer reveals conserved disease mechanism.PLoS One. 2022 Dec 12;17(12):e0278058. doi: 10.1371/journal.pone.0278058. eCollection 2022. PLoS One. 2022. PMID: 36508448 Free PMC article.
-
A novel diG motif in ORF3a protein of SARS-Cov-2 for intracellular transport.Front Cell Dev Biol. 2022 Nov 23;10:1011221. doi: 10.3389/fcell.2022.1011221. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506095 Free PMC article.
-
SARS-CoV-2 Nsp6 damages Drosophila heart and mouse cardiomyocytes through MGA/MAX complex-mediated increased glycolysis.Commun Biol. 2022 Sep 30;5(1):1039. doi: 10.1038/s42003-022-03986-6. Commun Biol. 2022. PMID: 36180527 Free PMC article.
-
Drosophila Innate Immunity Involves Multiple Signaling Pathways and Coordinated Communication Between Different Tissues.Front Immunol. 2022 Jul 7;13:905370. doi: 10.3389/fimmu.2022.905370. eCollection 2022. Front Immunol. 2022. PMID: 35911716 Free PMC article. Review.
References
-
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
-
- Singh A.D., Gupta D., Jameel S. Bioinformatic analysis of the SARS virus X1 protein shows it to be a calcium-binding protein. Curr. Sci. 2004;86:842–844.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

