SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II
- PMID: 16204216
- PMCID: PMC1456244
- DOI: 10.1534/genetics.105.047167
SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II
Abstract
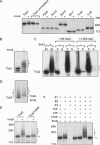
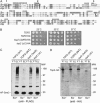
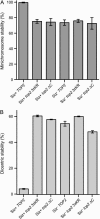
The Smt3 (SUMO) protein is conjugated to substrate proteins through a cascade of E1, E2, and E3 enzymes. In budding yeast, the E3 step in sumoylation is largely controlled by Siz1p and Siz2p. Analysis of Siz- cells shows that SUMO E3 is required for minichromosome segregation and thus has a positive role in maintaining the fidelity of mitotic transmission of genetic information. Sumoylation of the carboxy-terminus of Top2p, a known SUMO target, is mediated by Siz1p and Siz2p both in vivo and in vitro. Sumoylation in vitro reveals that Top2p is an extremely potent substrate for Smt3p conjugation and that chromatin-bound Top2p can still be sumoylated, unlike many other SUMO substrates. By combining mutations in the TOP2 sumoylation sites and the SIZ1 and SIZ2 genes we demonstrate that the minichromosome segregation defect and dicentric minichromosome stabilization, both characteristic for Smt3p-E3-deficient cells, are mediated by the lack of Top2p sumoylation in these cells. A role for Smt3p-modification as a signal for Top2p targeting to pericentromeric regions was suggested by an analysis of Top2p-Smt3p fusion. We propose a model for the positive control of the centromeric pool of Top2p, required for high segregation fidelity, by Smt3p modification.
Figures
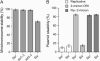
Similar articles
-
Cooperation of sumoylated chromosomal proteins in rDNA maintenance.PLoS Genet. 2008 Oct;4(10):e1000215. doi: 10.1371/journal.pgen.1000215. Epub 2008 Oct 10. PLoS Genet. 2008. PMID: 18846224 Free PMC article.
-
Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro.J Biochem. 2003 Apr;133(4):415-22. doi: 10.1093/jb/mvg054. J Biochem. 2003. PMID: 12761287
-
Multiple domains in Siz SUMO ligases contribute to substrate selectivity.J Cell Sci. 2006 Nov 15;119(Pt 22):4749-57. doi: 10.1242/jcs.03243. Epub 2006 Oct 31. J Cell Sci. 2006. PMID: 17077124
-
SUMOylation in control of accurate chromosome segregation during mitosis.Curr Protein Pept Sci. 2012 Aug;13(5):467-81. doi: 10.2174/138920312802430563. Curr Protein Pept Sci. 2012. PMID: 22812528 Free PMC article. Review.
-
SUMO conjugation - a mechanistic view.Biomol Concepts. 2017 Mar 1;8(1):13-36. doi: 10.1515/bmc-2016-0030. Biomol Concepts. 2017. PMID: 28284030 Review.
Cited by
-
Distinct functional domains of Ubc9 dictate cell survival and resistance to genotoxic stress.Mol Cell Biol. 2006 Jul;26(13):4958-69. doi: 10.1128/MCB.00160-06. Mol Cell Biol. 2006. PMID: 16782883 Free PMC article.
-
Topoisomerase I-dependent viability loss in saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair.Genetics. 2007 Sep;177(1):17-30. doi: 10.1534/genetics.107.074708. Epub 2007 Jul 1. Genetics. 2007. PMID: 17603101 Free PMC article.
-
Cooperation of sumoylated chromosomal proteins in rDNA maintenance.PLoS Genet. 2008 Oct;4(10):e1000215. doi: 10.1371/journal.pgen.1000215. Epub 2008 Oct 10. PLoS Genet. 2008. PMID: 18846224 Free PMC article.
-
The role of SUMOylation during development.Biochem Soc Trans. 2020 Apr 29;48(2):463-478. doi: 10.1042/BST20190390. Biochem Soc Trans. 2020. PMID: 32311032 Free PMC article. Review.
-
The SUMO Pathway in Mitosis.Adv Exp Med Biol. 2017;963:171-184. doi: 10.1007/978-3-319-50044-7_10. Adv Exp Med Biol. 2017. PMID: 28197912 Free PMC article. Review.
References
-
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner and S. J. Elledge, 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9: 1169–1182. - PubMed
-
- Champoux, J. J., 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70: 369–413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

