Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase
- PMID: 16189012
- PMCID: PMC1235819
- DOI: 10.1128/JVI.79.20.13105-13115.2005
Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase
Abstract
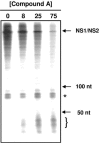
Respiratory syncytial virus (RSV) is a major cause of respiratory illness in infants, immunocompromised patients, and the elderly. New antiviral agents would be important tools in the treatment of acute RSV disease. RSV encodes its own RNA-dependent RNA polymerase that is responsible for the synthesis of both genomic RNA and subgenomic mRNAs. The viral polymerase also cotranscriptionally caps and polyadenylates the RSV mRNAs at their 5' and 3' ends, respectively. We have previously reported the discovery of the first nonnucleoside transcriptase inhibitor of RSV polymerase through high-throughput screening. Here we report the design of inhibitors that have improved potency both in vitro and in antiviral assays and that also exhibit activity in a mouse model of RSV infection. We have isolated virus with reduced susceptibility to this class of inhibitors. The mutations conferring resistance mapped to a novel motif within the RSV L gene, which encodes the catalytic subunit of RSV polymerase. This motif is distinct from the catalytic region of the L protein and bears some similarity to the nucleotide binding domain within nucleoside diphosphate kinases. These findings lead to the hypothesis that this class of inhibitors may block synthesis of RSV mRNAs by inhibiting guanylylation of viral transcripts. We show that short transcripts produced in the presence of inhibitor in vitro do not contain a 5' cap but, instead, are triphosphorylated, confirming this hypothesis. These inhibitors constitute useful tools for elucidating the molecular mechanism of RSV capping and represent valid leads for the development of novel anti-RSV therapeutics.
Figures


Similar articles
-
Respiratory Syncytial Virus Inhibitor AZ-27 Differentially Inhibits Different Polymerase Activities at the Promoter.J Virol. 2015 Aug;89(15):7786-98. doi: 10.1128/JVI.00530-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995255 Free PMC article.
-
RNA elongation by respiratory syncytial virus polymerase is calibrated by conserved region V.PLoS Pathog. 2017 Dec 27;13(12):e1006803. doi: 10.1371/journal.ppat.1006803. eCollection 2017 Dec. PLoS Pathog. 2017. PMID: 29281742 Free PMC article.
-
Discovery of 4'-chloromethyl-2'-deoxy-3',5'-di-O-isobutyryl-2'-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection.J Med Chem. 2015 Feb 26;58(4):1862-78. doi: 10.1021/jm5017279. Epub 2015 Feb 10. J Med Chem. 2015. PMID: 25667954
-
New antiviral approaches for respiratory syncytial virus and other mononegaviruses: Inhibiting the RNA polymerase.Antiviral Res. 2016 Oct;134:63-76. doi: 10.1016/j.antiviral.2016.08.006. Epub 2016 Aug 27. Antiviral Res. 2016. PMID: 27575793 Review.
-
2-5A antisense treatment of respiratory syncytial virus.Curr Opin Pharmacol. 2005 Oct;5(5):502-7. doi: 10.1016/j.coph.2005.05.003. Curr Opin Pharmacol. 2005. PMID: 16081320 Review.
Cited by
-
Intervention strategies for emerging viruses: use of antivirals.Curr Opin Virol. 2013 Apr;3(2):217-24. doi: 10.1016/j.coviro.2013.03.001. Epub 2013 Apr 4. Curr Opin Virol. 2013. PMID: 23562753 Free PMC article. Review.
-
Antiviral Efficacy of a Respiratory Syncytial Virus (RSV) Fusion Inhibitor in a Bovine Model of RSV Infection.Antimicrob Agents Chemother. 2015 Aug;59(8):4889-900. doi: 10.1128/AAC.00487-15. Epub 2015 Jun 8. Antimicrob Agents Chemother. 2015. PMID: 26055364 Free PMC article. Clinical Trial.
-
Viral replicons as valuable tools for drug discovery.Drug Discov Today. 2020 Jun;25(6):1026-1033. doi: 10.1016/j.drudis.2020.03.010. Epub 2020 Apr 6. Drug Discov Today. 2020. PMID: 32272194 Free PMC article. Review.
-
Ebola Virus Produces Discrete Small Noncoding RNAs Independently of the Host MicroRNA Pathway Which Lack RNA Interference Activity in Bat and Human Cells.J Virol. 2020 Feb 28;94(6):e01441-19. doi: 10.1128/JVI.01441-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31852785 Free PMC article.
-
Inhibition of a metal-dependent viral RNA triphosphatase by decavanadate.Biochem J. 2006 Sep 15;398(3):557-67. doi: 10.1042/BJ20060198. Biochem J. 2006. PMID: 16761952 Free PMC article.
References
-
- Austel, A., E. Kutter, U. Heider, W. Eberlein, W. Korginger, C. Lillie, W. Diederen, and W. Haarmann. November 1979. Imidazoisoquinoline-diones and salts thereof. U.S. Patent 4,176,184.
-
- Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. - PubMed
-
- Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. - PubMed
-
- Barik, S. 1993. The structure of the 5′ terminal cap of the respiratory syncytial virus mRNA. J. Gen. Virol. 74:485-490. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

