Why highly expressed proteins evolve slowly
- PMID: 16176987
- PMCID: PMC1242296
- DOI: 10.1073/pnas.0504070102
Why highly expressed proteins evolve slowly
Abstract
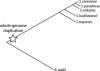
Much recent work has explored molecular and population-genetic constraints on the rate of protein sequence evolution. The best predictor of evolutionary rate is expression level, for reasons that have remained unexplained. Here, we hypothesize that selection to reduce the burden of protein misfolding will favor protein sequences with increased robustness to translational missense errors. Pressure for translational robustness increases with expression level and constrains sequence evolution. Using several sequenced yeast genomes, global expression and protein abundance data, and sets of paralogs traceable to an ancient whole-genome duplication in yeast, we rule out several confounding effects and show that expression level explains roughly half the variation in Saccharomyces cerevisiae protein evolutionary rates. We examine causes for expression's dominant role and find that genome-wide tests favor the translational robustness explanation over existing hypotheses that invoke constraints on function or translational efficiency. Our results suggest that proteins evolve at rates largely unrelated to their functions and can explain why highly expressed proteins evolve slowly across the tree of life.
Figures
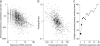
Similar articles
-
Population genetics of translational robustness.Genetics. 2006 May;173(1):473-81. doi: 10.1534/genetics.105.051300. Epub 2006 Feb 19. Genetics. 2006. PMID: 16489231 Free PMC article.
-
Retention of protein complex membership by ancient duplicated gene products in budding yeast.Trends Genet. 2007 Jun;23(6):266-9. doi: 10.1016/j.tig.2007.03.012. Epub 2007 Apr 10. Trends Genet. 2007. PMID: 17428571 Review.
-
Rate of protein evolution versus fitness effect of gene deletion.Mol Biol Evol. 2003 May;20(5):772-4. doi: 10.1093/molbev/msg078. Epub 2003 Apr 2. Mol Biol Evol. 2003. PMID: 12679525
-
Evolutionary dynamics and functional specialization of plant paralogs formed by whole and small-scale genome duplications.Mol Biol Evol. 2012 Nov;29(11):3541-51. doi: 10.1093/molbev/mss162. Epub 2012 Jun 24. Mol Biol Evol. 2012. PMID: 22734049
-
Evolutionary genomics: yeasts accelerate beyond BLAST.Curr Biol. 2004 May 25;14(10):R392-4. doi: 10.1016/j.cub.2004.05.015. Curr Biol. 2004. PMID: 15186766 Review.
Cited by
-
Molecular evolution of anthocyanin pigmentation genes following losses of flower color.BMC Evol Biol. 2016 May 10;16:98. doi: 10.1186/s12862-016-0675-3. BMC Evol Biol. 2016. PMID: 27161359 Free PMC article.
-
Population-level transposable element expression dynamics influence trait evolution in a fungal crop pathogen.mBio. 2024 Mar 13;15(3):e0284023. doi: 10.1128/mbio.02840-23. Epub 2024 Feb 13. mBio. 2024. PMID: 38349152 Free PMC article.
-
Gene Coexpression Network Reveals Highly Conserved, Well-Regulated Anti-Ageing Mechanisms in Old Ant Queens.Genome Biol Evol. 2021 Jun 8;13(6):evab093. doi: 10.1093/gbe/evab093. Genome Biol Evol. 2021. PMID: 33944936 Free PMC article.
-
The Phenotypic Plasticity of Duplicated Genes in Saccharomyces cerevisiae and the Origin of Adaptations.G3 (Bethesda). 2017 Jan 5;7(1):63-75. doi: 10.1534/g3.116.035329. G3 (Bethesda). 2017. PMID: 27799339 Free PMC article.
-
Contribution of the epigenetic mark H3K27me3 to functional divergence after whole genome duplication in Arabidopsis.Genome Biol. 2012 Oct 3;13(10):R94. doi: 10.1186/gb-2012-13-10-r94. Genome Biol. 2012. PMID: 23034476 Free PMC article.
References
-
- Wall, D. P., Fraser, H. B. & Hirsh, A. E. (2003) Bioinformatics 19, 1710-1711. - PubMed
-
- Graur, D. & Li, W.-H. (2000) Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA).
-
- Zuckerkandl, E. (1976) J. Mol. Evol. 7, 167-183. - PubMed
-
- Fraser, H. B., Hirsh, A. E., Steinmetz, L. M., Scharfe, C. & Feldman, M. W. (2002) Science 296, 750-752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

