Systematic survey reveals general applicability of "guilt-by-association" within gene coexpression networks
- PMID: 16162296
- PMCID: PMC1239911
- DOI: 10.1186/1471-2105-6-227
Systematic survey reveals general applicability of "guilt-by-association" within gene coexpression networks
Abstract
Background: Biological processes are carried out by coordinated modules of interacting molecules. As clustering methods demonstrate that genes with similar expression display increased likelihood of being associated with a common functional module, networks of coexpressed genes provide one framework for assigning gene function. This has informed the guilt-by-association (GBA) heuristic, widely invoked in functional genomics. Yet although the idea of GBA is accepted, the breadth of GBA applicability is uncertain.
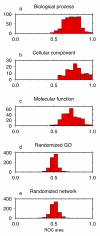
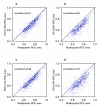
Results: We developed methods to systematically explore the breadth of GBA across a large and varied corpus of expression data to answer the following question: To what extent is the GBA heuristic broadly applicable to the transcriptome and conversely how broadly is GBA captured by a priori knowledge represented in the Gene Ontology (GO)? Our study provides an investigation of the functional organization of five coexpression networks using data from three mammalian organisms. Our method calculates a probabilistic score between each gene and each Gene Ontology category that reflects coexpression enrichment of a GO module. For each GO category we use Receiver Operating Curves to assess whether these probabilistic scores reflect GBA. This methodology applied to five different coexpression networks demonstrates that the signature of guilt-by-association is ubiquitous and reproducible and that the GBA heuristic is broadly applicable across the population of nine hundred Gene Ontology categories. We also demonstrate the existence of highly reproducible patterns of coexpression between some pairs of GO categories.
Conclusion: We conclude that GBA has universal value and that transcriptional control may be more modular than previously realized. Our analyses also suggest that methodologies combining coexpression measurements across multiple genes in a biologically-defined module can aid in characterizing gene function or in characterizing whether pairs of functions operate together.
Figures


Similar articles
-
Proteome Profiling Outperforms Transcriptome Profiling for Coexpression Based Gene Function Prediction.Mol Cell Proteomics. 2017 Jan;16(1):121-134. doi: 10.1074/mcp.M116.060301. Epub 2016 Nov 11. Mol Cell Proteomics. 2017. PMID: 27836980 Free PMC article.
-
Network‑based gene function inference method to predict optimal gene functions associated with fetal growth restriction.Mol Med Rep. 2018 Sep;18(3):3003-3010. doi: 10.3892/mmr.2018.9232. Epub 2018 Jun 29. Mol Med Rep. 2018. PMID: 30015878
-
Assessment and integration of publicly available SAGE, cDNA microarray, and oligonucleotide microarray expression data for global coexpression analyses.Genomics. 2005 Oct;86(4):476-88. doi: 10.1016/j.ygeno.2005.06.009. Genomics. 2005. PMID: 16098712
-
Identification of a gene module associated with BMD through the integration of network analysis and genome-wide association data.J Bone Miner Res. 2010 Nov;25(11):2359-67. doi: 10.1002/jbmr.138. J Bone Miner Res. 2010. PMID: 20499364
-
Comparative co-expression analysis in plant biology.Plant Cell Environ. 2012 Oct;35(10):1787-98. doi: 10.1111/j.1365-3040.2012.02517.x. Epub 2012 May 10. Plant Cell Environ. 2012. PMID: 22489681 Review.
Cited by
-
A graph theoretical approach to experimental prioritization in genome-scale investigations.Mamm Genome. 2024 Dec;35(4):724-733. doi: 10.1007/s00335-024-10066-z. Epub 2024 Aug 27. Mamm Genome. 2024. PMID: 39191873 Free PMC article.
-
Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders.Nat Rev Genet. 2015 Aug;16(8):441-58. doi: 10.1038/nrg3934. Epub 2015 Jul 7. Nat Rev Genet. 2015. PMID: 26149713 Free PMC article. Review.
-
A sparse Bayesian factor model for the construction of gene co-expression networks from single-cell RNA sequencing count data.BMC Bioinformatics. 2020 Aug 18;21(1):361. doi: 10.1186/s12859-020-03707-y. BMC Bioinformatics. 2020. PMID: 32811424 Free PMC article.
-
Blood-derived microRNAs are related to cognitive domains in the general population.Alzheimers Dement. 2024 Oct;20(10):7138-7159. doi: 10.1002/alz.14197. Epub 2024 Aug 29. Alzheimers Dement. 2024. PMID: 39210637 Free PMC article.
-
Network-based inference from complex proteomic mixtures using SNIPE.Bioinformatics. 2012 Dec 1;28(23):3115-22. doi: 10.1093/bioinformatics/bts594. Epub 2012 Oct 11. Bioinformatics. 2012. PMID: 23060611 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

