Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing
- PMID: 16141067
- PMCID: PMC1850959
- DOI: 10.1126/science.1113150
Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing
Abstract
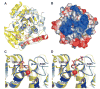
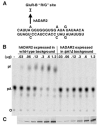
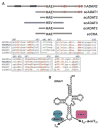
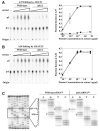
We report the crystal structure of the catalytic domain of human ADAR2, an RNA editing enzyme, at 1.7 angstrom resolution. The structure reveals a zinc ion in the active site and suggests how the substrate adenosine is recognized. Unexpectedly, inositol hexakisphosphate (IP6) is buried within the enzyme core, contributing to the protein fold. Although there are no reports that adenosine deaminases that act on RNA (ADARs) require a cofactor, we show that IP6 is required for activity. Amino acids that coordinate IP6 in the crystal structure are conserved in some adenosine deaminases that act on transfer RNA (tRNA) (ADATs), related enzymes that edit tRNA. Indeed, IP6 is also essential for in vivo and in vitro deamination of adenosine 37 of tRNAala by ADAT1.
Figures


Similar articles
-
The properties of a tRNA-specific adenosine deaminase from Drosophila melanogaster support an evolutionary link between pre-mRNA editing and tRNA modification.Mol Cell Biol. 2000 Feb;20(3):825-33. doi: 10.1128/MCB.20.3.825-833.2000. Mol Cell Biol. 2000. PMID: 10629039 Free PMC article.
-
How do ADARs bind RNA? New protein-RNA structures illuminate substrate recognition by the RNA editing ADARs.Bioessays. 2017 Apr;39(4):10.1002/bies.201600187. doi: 10.1002/bies.201600187. Epub 2017 Feb 20. Bioessays. 2017. PMID: 28217931 Free PMC article. Review.
-
Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities.Biochemistry. 2000 Oct 24;39(42):12875-84. doi: 10.1021/bi001383g. Biochemistry. 2000. PMID: 11041852
-
Mechanistic insights into editing-site specificity of ADARs.Proc Natl Acad Sci U S A. 2012 Nov 27;109(48):E3295-304. doi: 10.1073/pnas.1212548109. Epub 2012 Nov 5. Proc Natl Acad Sci U S A. 2012. PMID: 23129636 Free PMC article.
-
RNA editing by base deamination: more enzymes, more targets, new mysteries.Trends Biochem Sci. 2001 Jun;26(6):376-84. doi: 10.1016/s0968-0004(01)01827-8. Trends Biochem Sci. 2001. PMID: 11406411 Review.
Cited by
-
ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells.Nat Commun. 2021 Mar 12;12(1):1654. doi: 10.1038/s41467-021-21921-x. Nat Commun. 2021. PMID: 33712600 Free PMC article.
-
Defining signal transduction by inositol phosphates.Subcell Biochem. 2012;59:389-412. doi: 10.1007/978-94-007-3015-1_13. Subcell Biochem. 2012. PMID: 22374098 Free PMC article. Review.
-
Structure of the TRPA1 ion channel suggests regulatory mechanisms.Nature. 2015 Apr 23;520(7548):511-7. doi: 10.1038/nature14367. Epub 2015 Apr 8. Nature. 2015. PMID: 25855297 Free PMC article.
-
Nucleoside analog studies indicate mechanistic differences between RNA-editing adenosine deaminases.Nucleic Acids Res. 2012 Oct;40(19):9825-35. doi: 10.1093/nar/gks752. Epub 2012 Aug 11. Nucleic Acids Res. 2012. PMID: 22885375 Free PMC article.
-
Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from maize: molecular and biochemical characterization.Plant Physiol. 2007 Jul;144(3):1278-91. doi: 10.1104/pp.107.095455. Epub 2007 May 25. Plant Physiol. 2007. PMID: 17535825 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

