Molecular interactions in the assembly of coronaviruses
- PMID: 16139595
- PMCID: PMC7112327
- DOI: 10.1016/S0065-3527(05)64006-7
Molecular interactions in the assembly of coronaviruses
Abstract
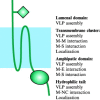
This chapter describes the interactions between the different structural components of the viruses and discusses their relevance for the process of virion formation. Two key factors determine the efficiency of the assembly process: intracellular transport and molecular interactions. Many viruses have evolved elaborate strategies to ensure the swift and accurate delivery of the virion components to the cellular compartment(s) where they must meet and form (sub) structures. Assembly of viruses starts in the nucleus by the encapsidation of viral DNA, using cytoplasmically synthesized capsid proteins; nucleocapsids then migrate to the cytosol, by budding at the inner nuclear membrane followed by deenvelopment, to pick up the tegument proteins.
Figures
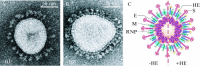
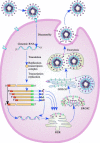

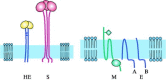
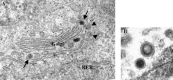

Similar articles
-
Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus.Virus Res. 2004 Oct;105(2):121-5. doi: 10.1016/j.virusres.2004.05.002. Virus Res. 2004. PMID: 15351485 Free PMC article.
-
The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles.J Virol. 2008 Nov;82(22):11318-30. doi: 10.1128/JVI.01052-08. Epub 2008 Aug 27. J Virol. 2008. PMID: 18753196 Free PMC article.
-
Peptide domain involved in the interaction between membrane protein and nucleocapsid protein of SARS-associated coronavirus.J Biochem Mol Biol. 2005 Jul 31;38(4):381-5. doi: 10.5483/bmbrep.2005.38.4.381. J Biochem Mol Biol. 2005. PMID: 16053703
-
An overall picture of SARS coronavirus (SARS-CoV) genome-encoded major proteins: structures, functions and drug development.Curr Pharm Des. 2006;12(35):4539-53. doi: 10.2174/138161206779010459. Curr Pharm Des. 2006. PMID: 17168760 Review.
-
Background paper. Coronavirus M and HE: two peculiar glycoproteins.Adv Exp Med Biol. 1990;276:91-4. doi: 10.1007/978-1-4684-5823-7_13. Adv Exp Med Biol. 1990. PMID: 2103107 Review. No abstract available.
Cited by
-
Calcium‐dependent antimicrobials: Nature‐inspired materials and designs.Exploration (Beijing). 2024 Mar 12;4(5):20230099. doi: 10.1002/EXP.20230099. eCollection 2024 Oct. Exploration (Beijing). 2024. PMID: 39439493 Free PMC article. Review. Catalan.
-
AlphaFold2 Reveals Structural Patterns of Seasonal Haplotype Diversification in SARS-CoV-2 Nucleocapsid Protein Variants.Viruses. 2024 Aug 25;16(9):1358. doi: 10.3390/v16091358. Viruses. 2024. PMID: 39339835 Free PMC article.
-
Co-Mutations and Possible Variation Tendency of the Spike RBD and Membrane Protein in SARS-CoV-2 by Machine Learning.Int J Mol Sci. 2024 Apr 25;25(9):4662. doi: 10.3390/ijms25094662. Int J Mol Sci. 2024. PMID: 38731879 Free PMC article.
-
Emerging Trends of Gold Nanostructures for Point-of-Care Biosensor-Based Detection of COVID-19.Mol Biotechnol. 2024 May 4. doi: 10.1007/s12033-024-01157-y. Online ahead of print. Mol Biotechnol. 2024. PMID: 38703305 Review.
-
Elucidating the behavior of the SARS-CoV-2 virus surface at vapor-liquid interfaces using molecular dynamics simulation.Proc Natl Acad Sci U S A. 2024 Mar 26;121(13):e2317194121. doi: 10.1073/pnas.2317194121. Epub 2024 Mar 19. Proc Natl Acad Sci U S A. 2024. PMID: 38502700 Free PMC article.
References
-
- Armstrong J., Patel S. The Golgi sorting domain of coronavirus E1 protein. J. Cell Sci. 1991;98:567–575. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

