The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene
- PMID: 16135788
- PMCID: PMC1234311
- DOI: 10.1128/MCB.25.18.7953-7965.2005
The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene
Abstract
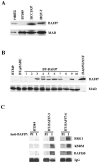
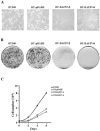
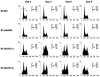
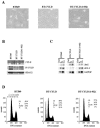
Mutation of BRG1, hBRM, and their associated factors, INI1 and BAF57, in primary human tumors has suggested that inactivation of human SWI/SNF (hSWI/SNF) complexes may be involved in neoplastic transformation. BT549 is an invasive human breast carcinoma cell line that lacks expression of BAF57, a key hSWI/SNF subunit that mediates interaction with transcriptional activators and corepressors. In this study we investigated the role of BAF57 in suppressing tumorigenesis by establishing BT549 stable cell lines that expresses full-length BAF57 protein. BT549 clones expressing BAF57 demonstrated marked phenotypic changes, slow growth kinetics, and restoration of contact inhibition. Altered growth was found to be due in part to cell cycle arrest and induction of apoptosis. Furthermore, microarray analysis revealed that BAF57-mediated cell death was associated with up-regulation of proapoptotic genes including the tumor suppressor familial cylindromatosis (CYLD), which was found to be a direct target of BAF57 as determined by chromatin immunoprecipitation analysis. Increased expression of CYLD in BT549 cells induced apoptosis, while its suppression by small interfering RNA inhibited cell death in BAF57 expressing BT549 cells. These findings demonstrate the importance of BAF57 in cell growth regulation and provide a novel link between hSWI/SNF chromatin remodelers and apoptosis.
Figures


Similar articles
-
Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes.Mol Cell Biol. 2004 Nov;24(21):9630-45. doi: 10.1128/MCB.24.21.9630-9645.2004. Mol Cell Biol. 2004. PMID: 15485929 Free PMC article.
-
Identification of BAF57 mutations in human breast cancer cell lines.Breast Cancer Res Treat. 2006 Jul;98(2):191-8. doi: 10.1007/s10549-005-9149-9. Epub 2006 Mar 15. Breast Cancer Res Treat. 2006. PMID: 16538531
-
A role for BAF57 in cell cycle-dependent transcriptional regulation by the SWI/SNF chromatin remodeling complex.Cancer Res. 2010 Jun 1;70(11):4402-11. doi: 10.1158/0008-5472.CAN-09-2767. Epub 2010 May 11. Cancer Res. 2010. PMID: 20460533 Free PMC article.
-
When the SWI/SNF complex remodels...the cell cycle.Oncogene. 2001 May 28;20(24):3067-75. doi: 10.1038/sj.onc.1204331. Oncogene. 2001. PMID: 11420722 Review.
-
The developmental and pathogenic roles of BAF57, a special subunit of the BAF chromatin-remodeling complex.FEBS Lett. 2016 Jun;590(11):1555-69. doi: 10.1002/1873-3468.12201. Epub 2016 May 20. FEBS Lett. 2016. PMID: 27149204 Review.
Cited by
-
Trisomy 19 ependymoma, a newly recognized genetico-histological association, including clear cell ependymoma.Mol Cancer. 2007 Jul 12;6:47. doi: 10.1186/1476-4598-6-47. Mol Cancer. 2007. PMID: 17626628 Free PMC article.
-
Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors.Mol Cell Biol. 2007 Jun;27(11):4018-27. doi: 10.1128/MCB.01839-06. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403896 Free PMC article.
-
The E3 ubiquitin ligase activity of Trip12 is essential for mouse embryogenesis.PLoS One. 2011;6(10):e25871. doi: 10.1371/journal.pone.0025871. Epub 2011 Oct 18. PLoS One. 2011. PMID: 22028794 Free PMC article.
-
Familial Syndromes Involving Meningiomas Provide Mechanistic Insight Into Sporadic Disease.Neurosurgery. 2018 Dec 1;83(6):1107-1118. doi: 10.1093/neuros/nyy121. Neurosurgery. 2018. PMID: 29660026 Free PMC article. Review.
-
ARID1B, a member of the human SWI/SNF chromatin remodeling complex, exhibits tumour-suppressor activities in pancreatic cancer cell lines.Br J Cancer. 2013 May 28;108(10):2056-62. doi: 10.1038/bjc.2013.200. Epub 2013 May 9. Br J Cancer. 2013. PMID: 23660946 Free PMC article.
References
-
- Abou El Hassan, M. A., D. C. Mastenbroek, W. R. Gerritsen, G. Giaccone, and F. A. Kruyt. 2004. Overexpression of Bcl2 abrogates chemo- and radiotherapy-induced sensitisation of NCI-H460 non-small-cell lung cancer cells to adenovirus-mediated expression of full-length TRAIL. Br. J. Cancer 91:171-177. - PMC - PubMed
-
- Ae, K., N. Kobayashi, R. Sakuma, T. Ogata, H. Kuroda, N. Kawaguchi, K. Shinomiya, and Y. Kitamura. 2002. Chromatin remodeling factor encoded by ini1 induces G1 arrest and apoptosis in ini1-deficient cells. Oncogene 21:3112-3120. - PubMed
-
- Baker, K. M., G. Wei, A. E. Schaffner, and M. C. Ostrowski. 2003. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J. Biol. Chem. 278:17876-17884. - PubMed
-
- Battaglioli, E., M. E. Andres, D. W. Rose, J. G. Chenoweth, M. G. Rosenfeld, M. E. Anderson, and G. Mandel. 2002. REST repression of neuronal genes requires components of the hSWI/SNF complex. J. Biol. Chem. 277:41038-41045. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
