Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice
- PMID: 16126864
- PMCID: PMC1203379
- DOI: 10.1104/pp.105.063420
Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice
Abstract
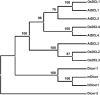
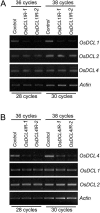
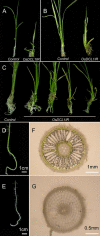
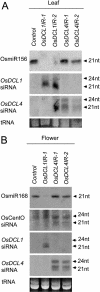
MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) are two types of noncoding RNAs involved in developmental regulation, genome maintenance, and defense in eukaryotes. The activity of Dicer or Dicer-like (DCL) proteins is required for the maturation of miRNAs and siRNAs. In this study, we cloned and sequenced 66 candidate rice (Oryza sativa) miRNAs out of 1,650 small RNA sequences (19 to approximately 25 nt), and they could be further grouped into 21 families, 12 of which are newly identified and three of which, OsmiR528, OsmiR529, and OsmiR530, have been confirmed by northern blot. To study the function of rice DCL proteins (OsDCLs) in the biogenesis of miRNAs and siRNAs, we searched genome databases and identified four OsDCLs. An RNA interference approach was applied to knock down two OsDCLs, OsDCL1 and OsDCL4, respectively. Strong loss of function of OsDCL1IR transformants that expressed inverted repeats of OsDCL1 resulted in developmental arrest at the seedling stage, and weak loss of function of OsDCL1IR transformants caused pleiotropic developmental defects. Moreover, all miRNAs tested were greatly reduced in OsDCL1IR but not OsDCL4IR transformants, indicating that OsDCL1 plays a critical role in miRNA processing in rice. In contrast, the production of siRNA from transgenic inverted repeats and endogenous CentO regions were not affected in either OsDCL1IR or OsDCL4IR transformants, suggesting that the production of miRNAs and siRNAs is via distinct OsDCLs.
Figures



Similar articles
-
Repression of microRNA biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice.Plant Sci. 2015 Aug;237:24-32. doi: 10.1016/j.plantsci.2015.05.002. Epub 2015 May 11. Plant Sci. 2015. PMID: 26089149
-
Roles of DCL4 and DCL3b in rice phased small RNA biogenesis.Plant J. 2012 Feb;69(3):462-74. doi: 10.1111/j.1365-313X.2011.04805.x. Epub 2011 Nov 23. Plant J. 2012. PMID: 21973320
-
Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways.Plant Cell. 2005 Nov;17(11):2873-85. doi: 10.1105/tpc.105.036608. Epub 2005 Oct 7. Plant Cell. 2005. PMID: 16214897 Free PMC article.
-
Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and Dicer-processing.J Plant Res. 2017 Jan;130(1):17-23. doi: 10.1007/s10265-016-0878-0. Epub 2016 Nov 29. J Plant Res. 2017. PMID: 27900550 Free PMC article. Review.
-
The expanding world of small RNAs in plants.Nat Rev Mol Cell Biol. 2015 Dec;16(12):727-41. doi: 10.1038/nrm4085. Epub 2015 Nov 4. Nat Rev Mol Cell Biol. 2015. PMID: 26530390 Free PMC article. Review.
Cited by
-
High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice.BMC Plant Biol. 2012 Aug 3;12:132. doi: 10.1186/1471-2229-12-132. BMC Plant Biol. 2012. PMID: 22862743 Free PMC article.
-
MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds.Sci Rep. 2016 Aug 30;6:32158. doi: 10.1038/srep32158. Sci Rep. 2016. PMID: 27574184 Free PMC article.
-
Small RNA profiling in two Brassica napus cultivars identifies microRNAs with oil production- and development-correlated expression and new small RNA classes.Plant Physiol. 2012 Feb;158(2):813-23. doi: 10.1104/pp.111.187666. Epub 2011 Dec 2. Plant Physiol. 2012. PMID: 22138974 Free PMC article.
-
Fine tuning of auxin signaling by miRNAs.Physiol Mol Biol Plants. 2008 Apr;14(1-2):81-90. doi: 10.1007/s12298-008-0007-1. Epub 2008 Jun 15. Physiol Mol Biol Plants. 2008. PMID: 23572875 Free PMC article.
-
Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses.Mol Genet Genomics. 2010 Dec;284(6):477-88. doi: 10.1007/s00438-010-0581-0. Epub 2010 Oct 13. Mol Genet Genomics. 2010. PMID: 20941508
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

