Loss of SMEK, a novel, conserved protein, suppresses MEK1 null cell polarity, chemotaxis, and gene expression defects
- PMID: 16107728
- PMCID: PMC1190274
- DOI: 10.1128/MCB.25.17.7839-7853.2005
Loss of SMEK, a novel, conserved protein, suppresses MEK1 null cell polarity, chemotaxis, and gene expression defects
Abstract
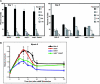
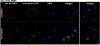
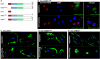
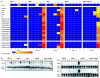
MEK/extracellular signal-regulated kinase (ERK) mitogen-activated protein kinase signaling is imperative for proper chemotaxis. Dictyostelium mek1(-) (MEK1 null) and erk1(-) cells exhibit severe defects in cell polarization and directional movement, but the molecules responsible for the mek1(-) and erk1(-) chemotaxis defects are unknown. Here, we describe a novel, evolutionarily conserved gene and protein (smkA and SMEK, respectively), whose loss partially suppresses the mek1(-) chemotaxis phenotypes. SMEK also has MEK1-independent functions: SMEK, but not MEK1, is required for proper cytokinesis during vegetative growth, timely exit from the mound stage during development, and myosin II assembly. SMEK localizes to the cell cortex through an EVH1 domain at its N terminus during vegetative growth. At the onset of development, SMEK translocates to the nucleus via a nuclear localization signal (NLS) at its C terminus. The importance of SMEK's nuclear localization is demonstrated by our findings that a mutant lacking the EVH1 domain complements SMEK deficiency, whereas a mutant lacking the NLS does not. Microarray analysis reveals that some genes are precociously expressed in mek1(-) and erk1(-) cells. The misexpression of some of these genes is suppressed in the smkA deletion. These data suggest that loss of MEK1/ERK1 signaling compromises gene expression and chemotaxis in a SMEK-dependent manner.
Figures



Similar articles
-
A hypothetical MEK1-MIP1-SMEK multiprotein signaling complex may function in Dictyostelium and mammalian cells.Int J Dev Biol. 2020;64(10-11-12):495-498. doi: 10.1387/ijdb.200140as. Int J Dev Biol. 2020. PMID: 33200810
-
Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis.Dev Cell. 2002 Jun;2(6):745-56. doi: 10.1016/s1534-5807(02)00186-7. Dev Cell. 2002. PMID: 12062087
-
Dictyostelium PAKc is required for proper chemotaxis.Mol Biol Cell. 2004 Dec;15(12):5456-69. doi: 10.1091/mbc.e04-04-0323. Epub 2004 Oct 13. Mol Biol Cell. 2004. PMID: 15483055 Free PMC article.
-
Signaling pathways controlling cell polarity and chemotaxis.Trends Biochem Sci. 2001 Sep;26(9):557-66. doi: 10.1016/s0968-0004(01)01934-x. Trends Biochem Sci. 2001. PMID: 11551793 Review.
-
Cell polarization: chemotaxis gets CRACKing.Curr Biol. 1999 Jan 28;9(2):R46-8. doi: 10.1016/s0960-9822(99)80007-4. Curr Biol. 1999. PMID: 10021352 Review.
Cited by
-
An integrated, cross-regulation pathway model involving activating/adaptive and feed-forward/feed-back loops for directed oscillatory cAMP signal-relay/response during the development of Dictyostelium.Front Cell Dev Biol. 2024 Jan 31;11:1263316. doi: 10.3389/fcell.2023.1263316. eCollection 2023. Front Cell Dev Biol. 2024. PMID: 38357530 Free PMC article.
-
Smek promotes histone deacetylation to suppress transcription of Wnt target gene brachyury in pluripotent embryonic stem cells.Cell Res. 2011 Jun;21(6):911-21. doi: 10.1038/cr.2011.47. Epub 2011 Mar 22. Cell Res. 2011. PMID: 21423269 Free PMC article.
-
Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes.Cell Mol Life Sci. 2014 Oct;71(19):3711-47. doi: 10.1007/s00018-014-1638-8. Epub 2014 May 21. Cell Mol Life Sci. 2014. PMID: 24846395 Free PMC article. Review.
-
Integration of diverse inputs in the regulation of Caenorhabditis elegans DAF-16/FOXO.Dev Dyn. 2010 May;239(5):1405-12. doi: 10.1002/dvdy.22244. Dev Dyn. 2010. PMID: 20140911 Free PMC article. Review.
-
The triglyceride catabolism regulated by a serine/threonine protein phosphatase, Smek1, is required for development and plant infection in Magnaporthe oryzae.Mol Plant Pathol. 2023 Oct;24(10):1256-1272. doi: 10.1111/mpp.13368. Epub 2023 Jun 26. Mol Plant Pathol. 2023. PMID: 37357820 Free PMC article.
References
-
- Anjard, C., F. Soderbom, and W. F. Loomis. 2001. Requirements for the adenylyl cyclases in the development of Dictyostelium. Development 128:3649-3654. - PubMed
-
- Aubry, L., and R. Firtel. 1999. Integration of signaling networks that regulate Dictyostelium differentiation. Annu. Rev. Cell Dev. Biol. 15:469-517. - PubMed
-
- Brahmbhatt, A. A., and R. L. Klemke. 2003. ERK and Rho differentially regulate pseudopodia growth and retraction during chemotaxis. J. Biol. Chem. 278:13016-13026. - PubMed
-
- Brakeman, P. R., A. A. Lanahan, R. O'Brien, K. Roche, C. A. Barnes, R. L. Huganir, and P. F. Worley. 1997. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature 386:284-288. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
