Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation
- PMID: 16107713
- PMCID: PMC1190292
- DOI: 10.1128/MCB.25.17.7665-7674.2005
Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation
Abstract
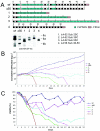
The C-terminal domain (CTD) of mammalian RNA polymerase II (Pol II) consists of 52 repeats of the consensus heptapeptide YSPTSPS and links transcription to the processing of pre-mRNA. The length of the CTD and the number of repeats diverging from the consensus sequence have increased through evolution, but their functional importance remains unknown. Here, we show that the deletion of repeats 1 to 3 or 52 leads to cleavage and degradation of the CTD from Pol II in vivo. Including these repeats, however, allowed the construction of stable, synthetic CTDs. To our surprise, polymerases consisting of just consensus repeats could support normal growth and viability of cells. We conclude that all other nonconsensus CTD repeats are dispensable for the transcription and pre-mRNA processing of genes essential for proliferation.
Figures



Similar articles
-
The C-Terminal Domain of RNA Polymerase II Is a Multivalent Targeting Sequence that Supports Drosophila Development with Only Consensus Heptads.Mol Cell. 2019 Mar 21;73(6):1232-1242.e4. doi: 10.1016/j.molcel.2019.01.008. Epub 2019 Feb 11. Mol Cell. 2019. PMID: 30765194 Free PMC article.
-
Growth retardation and neonatal lethality in mice with a homozygous deletion in the C-terminal domain of RNA polymerase II.Mol Gen Genet. 1999 Feb;261(1):100-5. doi: 10.1007/s004380050946. Mol Gen Genet. 1999. PMID: 10071215
-
Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3'-end cleavage.RNA. 2004 Apr;10(4):581-9. doi: 10.1261/rna.5207204. RNA. 2004. PMID: 15037767 Free PMC article.
-
Molecular evolution of the RNA polymerase II CTD.Trends Genet. 2008 Jun;24(6):289-96. doi: 10.1016/j.tig.2008.03.010. Epub 2008 May 9. Trends Genet. 2008. PMID: 18472177 Review.
-
Updating the RNA polymerase CTD code: adding gene-specific layers.Trends Genet. 2012 Jul;28(7):333-41. doi: 10.1016/j.tig.2012.03.007. Epub 2012 May 21. Trends Genet. 2012. PMID: 22622228 Review.
Cited by
-
Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3' end of human genes.Nat Struct Mol Biol. 2011 Sep 4;18(10):1115-23. doi: 10.1038/nsmb.2124. Nat Struct Mol Biol. 2011. PMID: 21892168
-
A single flexible RNAPII-CTD integrates many different transcriptional programs.Transcription. 2016 Mar 14;7(2):50-6. doi: 10.1080/21541264.2016.1163451. Epub 2016 Mar 17. Transcription. 2016. PMID: 26985717 Free PMC article.
-
Evolution of lysine acetylation in the RNA polymerase II C-terminal domain.BMC Evol Biol. 2015 Mar 10;15:35. doi: 10.1186/s12862-015-0327-z. BMC Evol Biol. 2015. PMID: 25887984 Free PMC article.
-
Analysis of RNA polymerase II ubiquitylation and proteasomal degradation.Methods. 2019 Apr 15;159-160:146-156. doi: 10.1016/j.ymeth.2019.02.005. Epub 2019 Feb 13. Methods. 2019. PMID: 30769100 Free PMC article.
-
Genetic analysis of the RNA polymerase II CTD in Drosophila.Methods. 2019 Apr 15;159-160:129-137. doi: 10.1016/j.ymeth.2019.01.010. Epub 2019 Jan 24. Methods. 2019. PMID: 30684537 Free PMC article. Review.
References
-
- Ares, M., Jr., and N. J. Proudfoot. 2005. The Spanish connection: transcription and mRNA processing get even closer. Cell 120:163-166. - PubMed
-
- Baskaran, R., S. R. Escobar, and J. Y. Wang. 1999. Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine (CTD)-tyrosine kinase-specific for the mammalian RNA polymerase II: possible role in transcription elongation. Cell Growth Differ. 10:387-396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
