microRNA target predictions across seven Drosophila species and comparison to mammalian targets
- PMID: 16103902
- PMCID: PMC1183519
- DOI: 10.1371/journal.pcbi.0010013
microRNA target predictions across seven Drosophila species and comparison to mammalian targets
Abstract
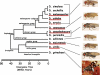
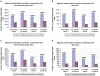
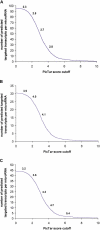
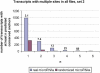
microRNAs are small noncoding genes that regulate the protein production of genes by binding to partially complementary sites in the mRNAs of targeted genes. Here, using our algorithm PicTar, we exploit cross-species comparisons to predict, on average, 54 targeted genes per microRNA above noise in Drosophila melanogaster. Analysis of the functional annotation of target genes furthermore suggests specific biological functions for many microRNAs. We also predict combinatorial targets for clustered microRNAs and find that some clustered microRNAs are likely to coordinately regulate target genes. Furthermore, we compare microRNA regulation between insects and vertebrates. We find that the widespread extent of gene regulation by microRNAs is comparable between flies and mammals but that certain microRNAs may function in clade-specific modes of gene regulation. One of these microRNAs (miR-210) is predicted to contribute to the regulation of fly oogenesis. We also list specific regulatory relationships that appear to be conserved between flies and mammals. Our findings provide the most extensive microRNA target predictions in Drosophila to date, suggest specific functional roles for most microRNAs, indicate the existence of coordinate gene regulation executed by clustered microRNAs, and shed light on the evolution of microRNA function across large evolutionary distances. All predictions are freely accessible at our searchable Web site http://pictar.bio.nyu.edu.
Conflict of interest statement
Figures


Similar articles
-
Combinatorial microRNA target predictions.Nat Genet. 2005 May;37(5):495-500. doi: 10.1038/ng1536. Epub 2005 Apr 3. Nat Genet. 2005. PMID: 15806104
-
Identification of Drosophila MicroRNA targets.PLoS Biol. 2003 Dec;1(3):E60. doi: 10.1371/journal.pbio.0000060. Epub 2003 Oct 13. PLoS Biol. 2003. PMID: 14691535 Free PMC article.
-
A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions.RNA. 2008 Nov;14(11):2290-6. doi: 10.1261/rna.1188208. Epub 2008 Sep 23. RNA. 2008. PMID: 18812437 Free PMC article.
-
The conserved miR-8/miR-200 microRNA family and their role in invertebrate and vertebrate neurogenesis.Cell Tissue Res. 2015 Jan;359(1):161-77. doi: 10.1007/s00441-014-1911-z. Epub 2014 May 30. Cell Tissue Res. 2015. PMID: 24875007 Review.
-
Insect microRNAs: Structure, function and evolution.Insect Biochem Mol Biol. 2007 Jan;37(1):3-9. doi: 10.1016/j.ibmb.2006.10.006. Epub 2006 Nov 7. Insect Biochem Mol Biol. 2007. PMID: 17175441 Review.
Cited by
-
MicroRNAs in inner ear biology and pathogenesis.Hear Res. 2012 May;287(1-2):6-14. doi: 10.1016/j.heares.2012.03.008. Epub 2012 Mar 29. Hear Res. 2012. PMID: 22484222 Free PMC article. Review.
-
Pervasive Selection against MicroRNA Target Sites in Human Populations.Mol Biol Evol. 2020 Dec 16;37(12):3399-3408. doi: 10.1093/molbev/msaa155. Mol Biol Evol. 2020. PMID: 32585012 Free PMC article.
-
Identification and characteristics of microRNAs from army worm, Spodoptera frugiperda cell line Sf21.PLoS One. 2015 Feb 18;10(2):e0116988. doi: 10.1371/journal.pone.0116988. eCollection 2015. PLoS One. 2015. PMID: 25693181 Free PMC article.
-
Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo.Dev Cell. 2012 Jul 17;23(1):202-9. doi: 10.1016/j.devcel.2012.05.013. Dev Cell. 2012. PMID: 22814608 Free PMC article.
-
Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway.RNA. 2010 Jan;16(1):43-56. doi: 10.1261/rna.1972910. Epub 2009 Nov 16. RNA. 2010. PMID: 19917635 Free PMC article.
References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

