Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects
- PMID: 16077009
- PMCID: PMC1182215
- DOI: 10.1101/gr.3771305
Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects
Abstract
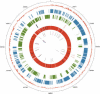
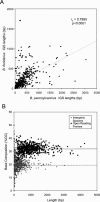
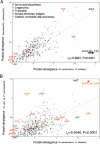
The distinct lifestyle of obligately intracellular bacteria can alter fundamental forces that drive and constrain genome change. In this study, sequencing the 792-kb genome of Blochmannia pennsylvanicus, an obligate endosymbiont of Camponotus pennsylvanicus, enabled us to trace evolutionary changes that occurred in the context of a bacterial-ant association. Comparison to the genome of Blochmannia floridanus reveals differential loss of genes involved in cofactor biosynthesis, the composition and structure of the cell wall and membrane, gene regulation, and DNA replication. However, the two Blochmannia species show complete conservation in the order and strand orientation of shared genes. This finding of extreme stasis in genome architecture, also reported previously for the aphid endosymbiont Buchnera, suggests that genome stability characterizes long-term bacterial mutualists of insects and constrains their evolutionary potential. Genome-wide analyses of protein divergences reveal 10- to 50-fold faster amino acid substitution rates in Blochmannia compared to related bacteria. Despite these varying features of genome evolution, a striking correlation in the relative divergences of proteins indicates parallel functional constraints on gene functions across ecologically distinct bacterial groups. Furthermore, the increased rates of amino acid substitution and gene loss in Blochmannia have occurred in a lineage-specific fashion, which may reflect life history differences of their ant hosts.
Figures



Similar articles
-
Unprecedented loss of ammonia assimilation capability in a urease-encoding bacterial mutualist.BMC Genomics. 2010 Dec 2;11:687. doi: 10.1186/1471-2164-11-687. BMC Genomics. 2010. PMID: 21126349 Free PMC article.
-
Host-symbiont stability and fast evolutionary rates in an ant-bacterium association: cospeciation of camponotus species and their endosymbionts, candidatus blochmannia.Syst Biol. 2004 Feb;53(1):95-110. doi: 10.1080/10635150490264842. Syst Biol. 2004. PMID: 14965905
-
Gene expression analysis of the endosymbiont-bearing midgut tissue during ontogeny of the carpenter ant Camponotus floridanus.J Insect Physiol. 2013 Jun;59(6):611-23. doi: 10.1016/j.jinsphys.2013.03.011. Epub 2013 Apr 6. J Insect Physiol. 2013. PMID: 23570961
-
Genome evolution in bacterial endosymbionts of insects.Nat Rev Genet. 2002 Nov;3(11):850-61. doi: 10.1038/nrg931. Nat Rev Genet. 2002. PMID: 12415315 Review.
-
Ancient bacterial endosymbionts of insects: Genomes as sources of insight and springboards for inquiry.Exp Cell Res. 2017 Sep 15;358(2):427-432. doi: 10.1016/j.yexcr.2017.04.028. Epub 2017 Apr 26. Exp Cell Res. 2017. PMID: 28454877 Review.
Cited by
-
Parallel genomic evolution and metabolic interdependence in an ancient symbiosis.Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19392-7. doi: 10.1073/pnas.0708855104. Epub 2007 Nov 28. Proc Natl Acad Sci U S A. 2007. PMID: 18048332 Free PMC article.
-
Role for Escherichia coli YidD in membrane protein insertion.J Bacteriol. 2011 Oct;193(19):5242-51. doi: 10.1128/JB.05429-11. Epub 2011 Jul 29. J Bacteriol. 2011. PMID: 21803992 Free PMC article.
-
Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture.Genome Biol Evol. 2011;3:209-18. doi: 10.1093/gbe/evr007. Epub 2011 Feb 2. Genome Biol Evol. 2011. PMID: 21292630 Free PMC article.
-
Cladogenesis and Genomic Streamlining in Extracellular Endosymbionts of Tropical Stink Bugs.Genome Biol Evol. 2018 Feb 1;10(2):680-693. doi: 10.1093/gbe/evy033. Genome Biol Evol. 2018. PMID: 29420776 Free PMC article.
-
Unprecedented loss of ammonia assimilation capability in a urease-encoding bacterial mutualist.BMC Genomics. 2010 Dec 2;11:687. doi: 10.1186/1471-2164-11-687. BMC Genomics. 2010. PMID: 21126349 Free PMC article.
References
-
- Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M., and Aksoy, S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32: 402–407. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Myers, W.E., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Andersson, B., Wentland, M.A., Ricafrente, J.Y., Liu, W., and Gibbs, R.A. 1996. A “double adaptor” method for improved shotgun library construction. Anal. Biochem. 236: 107–113. - PubMed
-
- Banerjee, T., Basak, S., Gupta, S.K., and Ghosh, T.C. 2004. Evolutionary forces in shaping the codon and amino acid usages in Blochmannia floridanus. J. Biomol. Struct. Dyn. 22: 13–24. - PubMed
-
- Baranov, P.V., Gesteland, R.F., and Atkins, J.F. 2002. Recoding: Translational bifurcations in gene expression. Gene 286: 187–201. - PubMed
Web site references
-
- http://ecocyc.org; Encyclopedia of Escherichia coli K12 Genes and Metabolism.
-
- http://genprotec.mbl.edu; GenProtEC E. coli genome and proteome database.
-
- http://selab.wustl.edu/cgi-bin/selab.pl?mode=software; tRNAscan-SE.
-
- http://www.genome.wi.mit.edu/wga; ARACHNE 2.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
