Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage
- PMID: 16061186
- PMCID: PMC4689305
- DOI: 10.1016/j.molcel.2005.07.011
Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage
Abstract
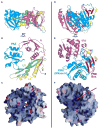
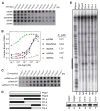
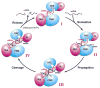
Argonaute (Ago) proteins constitute a key component of the RNA-induced silencing complex (RISC). We report the crystal structure of Aquifex aeolicus Ago (Aa-Ago) together with binding and cleavage studies, which establish this eubacterial Ago as a bona fide guide DNA strand-mediated site-specific RNA endonuclease. We have generated a stereochemically robust model of the complex, where the guide DNA-mRNA duplex is positioned within a basic channel spanning the bilobal interface, such that the 5' phosphate of the guide strand can be anchored in a basic pocket, and the mRNA can be positioned for site-specific cleavage by RNase H-type divalent cation-coordinated catalytic Asp residues of the PIWI domain. Domain swap experiments involving chimeras of human Ago (hAgo1) and cleavage-competent hAgo2 reinforce the role of the PIWI domain in "slicer" activity. We propose a four-step Ago-mediated catalytic cleavage cycle model, which provides distinct perspectives into the mechanism of guide strand-mediated mRNA cleavage within the RISC.
Figures




Similar articles
-
Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein.Nature. 2005 Mar 31;434(7033):666-70. doi: 10.1038/nature03514. Nature. 2005. PMID: 15800629 Free PMC article.
-
Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function.J Biol Chem. 2007 May 4;282(18):13824-32. doi: 10.1074/jbc.M608619200. Epub 2006 Nov 27. J Biol Chem. 2007. PMID: 17130125
-
A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA.Structure. 2006 Oct;14(10):1557-65. doi: 10.1016/j.str.2006.08.009. Structure. 2006. PMID: 17027504 Free PMC article.
-
When Argonaute takes out the ribonuclease sword.J Biol Chem. 2024 Jan;300(1):105499. doi: 10.1016/j.jbc.2023.105499. Epub 2023 Nov 27. J Biol Chem. 2024. PMID: 38029964 Free PMC article. Review.
-
The Argonautes.Cold Spring Harb Symp Quant Biol. 2006;71:67-72. doi: 10.1101/sqb.2006.71.048. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381282 Review.
Cited by
-
Key Mechanistic Principles and Considerations Concerning RNA Interference.Front Plant Sci. 2020 Aug 13;11:1237. doi: 10.3389/fpls.2020.01237. eCollection 2020. Front Plant Sci. 2020. PMID: 32903622 Free PMC article. Review.
-
Study on the clinical significance of Argonaute2 expression in colonic carcinoma by tissue microarray.Int J Clin Exp Pathol. 2013;6(3):476-84. Epub 2013 Feb 15. Int J Clin Exp Pathol. 2013. PMID: 23411422 Free PMC article.
-
Prokaryotic Argonaute Proteins as a Tool for Biotechnology.Mol Biol. 2022;56(6):854-873. doi: 10.1134/S0026893322060103. Epub 2022 Aug 30. Mol Biol. 2022. PMID: 36060308 Free PMC article.
-
Genomic identification, rapid evolution, and expression of Argonaute genes in the tilapia, Oreochromis niloticus.Dev Genes Evol. 2016 Sep;226(5):339-48. doi: 10.1007/s00427-016-0554-3. Epub 2016 Aug 5. Dev Genes Evol. 2016. PMID: 27491892
-
E-cadherin is transcriptionally activated via suppression of ZEB1 transcriptional repressor by small RNA-mediated gene silencing.PLoS One. 2011;6(12):e28688. doi: 10.1371/journal.pone.0028688. Epub 2011 Dec 21. PLoS One. 2011. PMID: 22205962 Free PMC article.
References
-
- Brunger AT. X-PLOR: A System for Crystallography and NMR, Version 3.1. New Haven, CT: Yale University Press; 1992.
-
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. - PubMed
-
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

