WW domains provide a platform for the assembly of multiprotein networks
- PMID: 16055720
- PMCID: PMC1190255
- DOI: 10.1128/MCB.25.16.7092-7106.2005
WW domains provide a platform for the assembly of multiprotein networks
Abstract
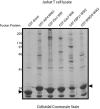
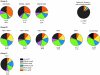
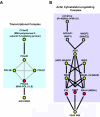
WW domains are protein modules that mediate protein-protein interactions through recognition of proline-rich peptide motifs and phosphorylated serine/threonine-proline sites. To pursue the functional properties of WW domains, we employed mass spectrometry to identify 148 proteins that associate with 10 human WW domains. Many of these proteins represent novel WW domain-binding partners and are components of multiprotein complexes involved in molecular processes, such as transcription, RNA processing, and cytoskeletal regulation. We validated one complex in detail, showing that WW domains of the AIP4 E3 protein-ubiquitin ligase bind directly to a PPXY motif in the p68 subunit of pre-mRNA cleavage and polyadenylation factor Im in a manner that promotes p68 ubiquitylation. The tested WW domains fall into three broad groups on the basis of hierarchical clustering with respect to their associated proteins; each such cluster of bound proteins displayed a distinct set of WW domain-binding motifs. We also found that separate WW domains from the same protein or closely related proteins can have different specificities for protein ligands and also demonstrated that a single polypeptide can bind multiple classes of WW domains through separate proline-rich motifs. These data suggest that WW domains provide a versatile platform to link individual proteins into physiologically important networks.
Figures






Similar articles
-
Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks.J Biol Chem. 2014 Mar 28;289(13):8865-80. doi: 10.1074/jbc.M113.506790. Epub 2014 Feb 18. J Biol Chem. 2014. PMID: 24550385 Free PMC article.
-
Solution structure and ligand recognition of the WW domain pair of the yeast splicing factor Prp40.J Mol Biol. 2002 Dec 6;324(4):807-22. doi: 10.1016/s0022-2836(02)01145-2. J Mol Biol. 2002. PMID: 12460579
-
Genome-wide analysis of the WW domain-containing protein genes in silkworm and their expansion in eukaryotes.Mol Genet Genomics. 2015 Jun;290(3):807-24. doi: 10.1007/s00438-014-0958-6. Epub 2014 Nov 26. Mol Genet Genomics. 2015. PMID: 25424044
-
WW domain-containing proteins: retrospectives and the future.Front Biosci (Landmark Ed). 2012 Jan 1;17(1):331-48. doi: 10.2741/3930. Front Biosci (Landmark Ed). 2012. PMID: 22201747 Review.
-
Recognition of proline-rich motifs by protein-protein-interaction domains.Angew Chem Int Ed Engl. 2005 May 6;44(19):2852-69. doi: 10.1002/anie.200400618. Angew Chem Int Ed Engl. 2005. PMID: 15880548 Review.
Cited by
-
Amot130 adapts atrophin-1 interacting protein 4 to inhibit yes-associated protein signaling and cell growth.J Biol Chem. 2013 May 24;288(21):15181-93. doi: 10.1074/jbc.M112.446534. Epub 2013 Apr 5. J Biol Chem. 2013. PMID: 23564455 Free PMC article.
-
Regulation of GPCR Trafficking by Ubiquitin.Prog Mol Biol Transl Sci. 2015;132:15-38. doi: 10.1016/bs.pmbts.2015.02.005. Epub 2015 Mar 25. Prog Mol Biol Transl Sci. 2015. PMID: 26055053 Free PMC article. Review.
-
Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains.EMBO Rep. 2006 Nov;7(11):1147-53. doi: 10.1038/sj.embor.7400822. Epub 2006 Oct 6. EMBO Rep. 2006. PMID: 17028573 Free PMC article.
-
Endogenous spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases AIP4 and AIP5.Biochem J. 2009 Sep 14;423(1):31-9. doi: 10.1042/BJ20082398. Biochem J. 2009. PMID: 19580544 Free PMC article.
-
Proline-rich sequence recognition: I. Marking GYF and WW domain assembly sites in early spliceosomal complexes.Mol Cell Proteomics. 2009 Nov;8(11):2461-73. doi: 10.1074/mcp.M900191-MCP200. Epub 2009 May 30. Mol Cell Proteomics. 2009. PMID: 19483244 Free PMC article.
References
-
- Bedford, M. T., D. Sarbassova, J. Xu, P. Leder, and M. B. Yaffe. 2000. A novel pro-Arg motif recognized by WW domains. J. Biol. Chem. 275:10359-10369. - PubMed
-
- Bednarek, A. K., K. J. Laflin, R. L. Daniel, Q. Liao, K. A. Hawkins, and C. M. Aldaz. 2000. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 60:2140-2145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
