Sensitive sequencing method for KRAS mutation detection by Pyrosequencing
- PMID: 16049314
- PMCID: PMC1867544
- DOI: 10.1016/S1525-1578(10)60571-5
Sensitive sequencing method for KRAS mutation detection by Pyrosequencing
Abstract
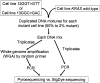
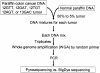
Both benign and malignant tumors represent heterogenous tissue containing tumor cells and non-neoplastic mesenchymal and inflammatory cells. To detect a minority of mutant KRAS alleles among abundant wild-type alleles, we developed a sensitive DNA sequencing assay using Pyrosequencing, ie, nucleotide extension sequencing with an allele quantification capability. We designed our Pyrosequencing assay for use with whole-genome-amplified DNA from paraffin-embedded tissue. Assessing various mixtures of DNA from mutant KRAS cell lines and DNA from a wild-type KRAS cell line, we found that mutation detection rates for Pyrosequencing were superior to dideoxy sequencing. In addition, Pyrosequencing proved superior to dideoxy sequencing in the detection of KRAS mutations from DNA mixtures of paraffin-embedded colon cancer and normal tissue as well as from paraffin-embedded pancreatic cancers. Quantification of mutant alleles by Pyrosequencing was precise and useful for assay validation, monitoring, and quality assurance. Our Pyrosequencing method is simple, robust, and sensitive, with a detection limit of approximately 5% mutant alleles. It is particularly useful for tumors containing abundant non-neoplastic cells. In addition, the applicability of this assay for DNA amplified by whole-genome amplification technique provides an expanded source of DNA for large-scale studies.
Figures




Similar articles
-
Pyrosequencing method to detect KRAS mutation in formalin-fixed and paraffin-embedded tumor tissues.Anal Biochem. 2009 Aug 15;391(2):166-8. doi: 10.1016/j.ab.2009.05.027. Epub 2009 May 21. Anal Biochem. 2009. PMID: 19464247
-
Polymerase chain reaction/sequencing analysis of ras mutations in paraffin-embedded tissues as compared with 3T3 transfection and polymerase chain reaction/sequencing of frozen tumor deoxyribonucleic acids.Lab Invest. 1992 Apr;66(4):504-11. Lab Invest. 1992. PMID: 1583889
-
KRAS detection in colonic tumors by DNA extraction from FTA paper: the molecular touch-prep.Diagn Mol Pathol. 2011 Dec;20(4):189-93. doi: 10.1097/PDM.0b013e318211d554. Diagn Mol Pathol. 2011. PMID: 22089345
-
Detection of K-ras mutations in pancreatic and hepatic neoplasms by non-isotopic mismatched polymerase chain reaction.Oncogene. 1991 May;6(5):857-62. Oncogene. 1991. PMID: 1646989
-
Development of pyrosequencing methods for the rapid detection of RAS mutations in clinical samples.Exp Mol Pathol. 2015 Oct;99(2):207-11. doi: 10.1016/j.yexmp.2015.07.003. Epub 2015 Jul 8. Exp Mol Pathol. 2015. PMID: 26163758
Cited by
-
Prognostic role of tumor PIK3CA mutation in colorectal cancer: a systematic review and meta-analysis.Ann Oncol. 2016 Oct;27(10):1836-48. doi: 10.1093/annonc/mdw264. Epub 2016 Jul 19. Ann Oncol. 2016. PMID: 27436848 Free PMC article. Review.
-
Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease.Mod Pathol. 2013 Apr;26(4):465-84. doi: 10.1038/modpathol.2012.214. Epub 2013 Jan 11. Mod Pathol. 2013. PMID: 23307060 Free PMC article. Review.
-
Detection of KRAS, NRAS and BRAF by mass spectrometry - a sensitive, reliable, fast and cost-effective technique.Diagn Pathol. 2015 Jul 30;10:132. doi: 10.1186/s13000-015-0364-3. Diagn Pathol. 2015. PMID: 26220423 Free PMC article.
-
Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication.J Natl Cancer Inst. 2013 Aug 7;105(15):1151-6. doi: 10.1093/jnci/djt173. Epub 2013 Jul 22. J Natl Cancer Inst. 2013. PMID: 23878352 Free PMC article.
-
Opportunities and challenges associated with clinical diagnostic genome sequencing: a report of the Association for Molecular Pathology.J Mol Diagn. 2012 Nov;14(6):525-40. doi: 10.1016/j.jmoldx.2012.04.006. Epub 2012 Aug 20. J Mol Diagn. 2012. PMID: 22918138 Free PMC article.
References
-
- Fakhrai-Rad H, Pourmand N, Ronaghi M. Pyrosequencing: an accurate detection platform for single nucleotide polymorphisms. Hum Mutat. 2002;19:479–485. - PubMed
-
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. - PubMed
-
- Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

