Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis
- PMID: 16027170
- PMCID: PMC1182342
- DOI: 10.1101/gad.1317105
Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis
Abstract
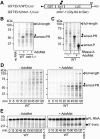
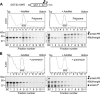
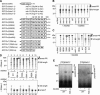
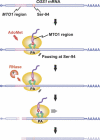
Expression of the Arabidopsis CGS1 gene that codes for cystathionine gamma-synthase is feedback regulated at the step of mRNA stability in response to S-adenosyl-L-methionine (AdoMet). A short stretch of amino acid sequence, called the MTO1 region, encoded by the first exon of CGS1 itself is involved in this regulation. Here, we demonstrate, using a cell-free system, that AdoMet induces temporal translation elongation arrest at the Ser-94 codon located immediately downstream of the MTO1 region, by analyzing a translation intermediate and performing primer extension inhibition (toeprint) analysis. This translation arrest precedes the formation of a degradation intermediate of CGS1 mRNA, which has its 5' end points near the 5' edge of the stalled ribosome. The position of ribosome stalling also suggests that the MTO1 region in nascent peptide resides in the ribosomal exit tunnel when translation elongation is temporarily arrested. In addition to the MTO1 region amino acid sequence, downstream Trp-93 is also important for the AdoMet-induced translation arrest. This is the first example of nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in eukaryotes. Furthermore, our data suggest that the ribosome stalls at the step of translocation rather than at the step of peptidyl transfer.
Figures



Similar articles
-
Nascent peptide-mediated translation elongation arrest of Arabidopsis thaliana CGS1 mRNA occurs autonomously.Plant Cell Physiol. 2008 Apr;49(4):549-56. doi: 10.1093/pcp/pcn028. Epub 2008 Feb 19. Plant Cell Physiol. 2008. PMID: 18285355
-
S-adenosyl-L-methionine induces compaction of nascent peptide chain inside the ribosomal exit tunnel upon translation arrest in the Arabidopsis CGS1 gene.J Biol Chem. 2011 Apr 29;286(17):14903-12. doi: 10.1074/jbc.M110.211656. Epub 2011 Feb 18. J Biol Chem. 2011. PMID: 21335553 Free PMC article.
-
Ribosome stacking defines CGS1 mRNA degradation sites during nascent peptide-mediated translation arrest.Plant Cell Physiol. 2008 Mar;49(3):314-23. doi: 10.1093/pcp/pcn005. Epub 2008 Jan 9. Plant Cell Physiol. 2008. PMID: 18184691
-
Arrest peptides: cis-acting modulators of translation.Annu Rev Biochem. 2013;82:171-202. doi: 10.1146/annurev-biochem-080211-105026. Annu Rev Biochem. 2013. PMID: 23746254 Review.
-
How Messenger RNA and Nascent Chain Sequences Regulate Translation Elongation.Annu Rev Biochem. 2018 Jun 20;87:421-449. doi: 10.1146/annurev-biochem-060815-014818. Annu Rev Biochem. 2018. PMID: 29925264 Free PMC article. Review.
Cited by
-
Surveillance pathways rescuing eukaryotic ribosomes lost in translation.Nat Rev Mol Cell Biol. 2012 Nov;13(11):727-35. doi: 10.1038/nrm3457. Epub 2012 Oct 17. Nat Rev Mol Cell Biol. 2012. PMID: 23072885 Review.
-
A systems view of the protein expression process.Syst Synth Biol. 2011 Dec;5(3-4):139-50. doi: 10.1007/s11693-011-9088-1. Epub 2011 Oct 19. Syst Synth Biol. 2011. PMID: 23205157 Free PMC article.
-
The Minimum Open Reading Frame, AUG-Stop, Induces Boron-Dependent Ribosome Stalling and mRNA Degradation.Plant Cell. 2016 Nov;28(11):2830-2849. doi: 10.1105/tpc.16.00481. Epub 2016 Oct 19. Plant Cell. 2016. PMID: 27760805 Free PMC article.
-
Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome.J Biol Chem. 2009 Apr 17;284(16):10343-52. doi: 10.1074/jbc.M808840200. Epub 2009 Feb 9. J Biol Chem. 2009. PMID: 19204001 Free PMC article.
-
Regulatory role of cystathionine-gamma-synthase and de novo synthesis of methionine in ethylene production during tomato fruit ripening.Plant Mol Biol. 2006 May;61(1-2):255-68. doi: 10.1007/s11103-006-0009-8. Plant Mol Biol. 2006. PMID: 16786305
References
-
- Anthony D.D. and Merrick, W.C. 1992. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J. Biol. Chem. 267: 1554-1562. - PubMed
-
- Bashan A., Zarivach, R., Schluenzen, F., Agmon, I., Harms, J., Auerbach, T., Baram, D., Berisio, R., Bartels, H., Hansen, H.A., et al. 2003. Ribosomal crystallography: Peptide bond formation and its inhibition. Biopolymers 70: 19-41. - PubMed
-
- Beier D., Stange, N., Gross, H.J., and Beier, H. 1991. Nuclear tRNA-Tyr genes are highly amplified at a single chromosomal site in the genome of Arabidopsis thaliana. Mol. Gen. Genet. 225: 72-80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
