Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation
- PMID: 16025112
- PMCID: PMC2659476
- DOI: 10.1038/ng1611
Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation
Abstract
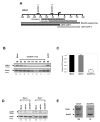
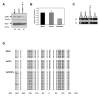
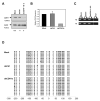
Double-stranded RNA molecules targeted to gene promoter regions can induce transcriptional gene silencing in a DNA cytosine methylation-dependent manner in plants (RNA-dependent DNA methylation). Whether a similar mechanism exists in mammalian systems is a vital and controversial issue. DNA methylation is an important component in mammalian gene silencing for normal processes such as gene imprinting and X-chromosome inactivation, and aberrant CpG island hypermethylation at tumor-suppressor promoters is associated with transcriptional silencing and loss of gene function in cancer. Hence, we investigated whether RNA-dependent DNA methylation might operate in human cancers to mediate epigenetic silencing using the endogenous gene CDH1 as a potential target. The loss of this cell-cell adhesion factor facilitates the metastatic process, and its promoter is frequently hypermethylated in breast and other cancers. We found that, although small double-stranded RNAs targeted exclusively to the CDH1 promoter could effectively induce transcriptional repression with chromatin changes characteristic of inactive promoters, this was entirely independent of DNA methylation. Moreover, we could accomplish such silencing in a cancer cell line genetically modified to lack virtually any capacity to methylate DNA.
Figures

Similar articles
-
Induction of DNA methylation and gene silencing by short interfering RNAs in human cells.Nature. 2004 Sep 9;431(7005):211-7. doi: 10.1038/nature02889. Epub 2004 Aug 15. Nature. 2004. Retraction in: Nature. 2006 Jun 29;441(7097):1176. doi: 10.1038/nature04952 PMID: 15311210 Retracted.
-
Transcriptional silencing and promoter methylation triggered by double-stranded RNA.EMBO J. 2000 Oct 2;19(19):5194-201. doi: 10.1093/emboj/19.19.5194. EMBO J. 2000. PMID: 11013221 Free PMC article.
-
Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers.Cancer Res. 2003 Jul 15;63(14):4174-80. Cancer Res. 2003. PMID: 12874023
-
Transcriptional gene silencing by short interfering RNAs.Curr Opin Mol Ther. 2005 Apr;7(2):125-31. Curr Opin Mol Ther. 2005. PMID: 15844619 Review.
-
DNA methylation and gene silencing in cancer: which is the guilty party?Oncogene. 2002 Aug 12;21(35):5380-7. doi: 10.1038/sj.onc.1205598. Oncogene. 2002. PMID: 12154400 Review.
Cited by
-
Small Interfering RNAs Targeting a Chromatin-Associated RNA Induce Its Transcriptional Silencing in Human Cells.Mol Cell Biol. 2022 Dec 15;42(12):e0027122. doi: 10.1128/mcb.00271-22. Epub 2022 Nov 29. Mol Cell Biol. 2022. PMID: 36445136 Free PMC article.
-
Emerging Contribution of PancRNAs in Cancer.Cancers (Basel). 2020 Jul 24;12(8):2035. doi: 10.3390/cancers12082035. Cancers (Basel). 2020. PMID: 32722129 Free PMC article. Review.
-
Small interfering RNA directed reversal of urokinase plasminogen activator demethylation inhibits prostate tumor growth and metastasis.Cancer Res. 2007 Jul 15;67(14):6637-46. doi: 10.1158/0008-5472.CAN-07-0751. Cancer Res. 2007. Retraction in: Cancer Res. 2018 Jun 15;78(12):3398. doi: 10.1158/0008-5472.CAN-18-1183 PMID: 17638874 Free PMC article. Retracted.
-
Quantitative analysis of conditional gene inactivation using rationally designed, tetracycline-controlled miRNAs.Nucleic Acids Res. 2010 Sep;38(17):e168. doi: 10.1093/nar/gkq616. Epub 2010 Jul 17. Nucleic Acids Res. 2010. PMID: 20639530 Free PMC article.
-
RNAi: a potential new class of therapeutic for human genetic disease.Hum Genet. 2011 Nov;130(5):583-605. doi: 10.1007/s00439-011-0995-8. Epub 2011 May 3. Hum Genet. 2011. PMID: 21537948 Review.
References
-
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–76. - PubMed
-
- Wassenegger M. RNA-directed DNA methylation. Plant Mol Biol. 2000;43:203–20. - PubMed
-
- Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–7. - PubMed
-
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

